miPEPs: Coding in non-coding plant primary transcripts of microRNAs

Most famous RNAs are coding or messenger RNAs (mRNAs) which are the ones that ribosomes read to synthesize proteins. However, many other RNAs are transcribed from DNA and are known as non-coding RNAs. Among those, ribosomic RNAs (rRNAs) which are part of the ribosomes and transfer RNAs (tRNAs) which deliver the amino acids to the ribosomes are widely known and studied from long time ago in High School biology courses. However, during the past two decades a much wider role has been evidenced for RNA through the discovery of many other types of non-coding RNAs (ncRNAs) with important regulatory functions. Today, we know the existence of, among others, siRNA, miRNA, lncRNA, piRNA, asRNA or tasiRNA.
For a long time the genomic DNA not leading to protein synthesis was considered as “junk DNA”. Although the existence of “junk DNA” is not completely disapproved, at present, it is getting clear that non-coding DNA plays important functions. A part from introns whose function in alternative splicing, process that leads to the generation of more than one mRNA from a single gene locus, is widely known, deep transcriptomic sequencing has revealed that at least 90% of the so-called DNA is transcribed into non-coding RNAs 1.
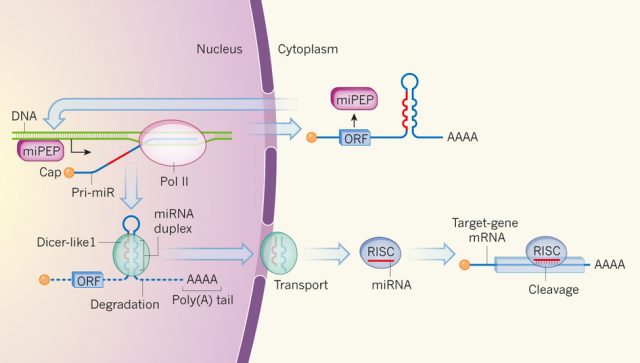
The most studied and relevant function of non-coding small RNAs was discovered late in the 90s and it is the post-transcriptional regulation of gene expression, one of the most important epigenetic mechanisms. In fact, a small RNA makes a double strand RNA with its complementary target mRNA and provokes its cleavage or reduced translation (Figure 1). That discovery has been an important breakthrough in gene expression regulation and opened an avenue for biotechnological applications including the known technique for gene silencing, RNA interference.
Briefly, miRNAs are intronic or intergenic regions which are transcribed by RNA polymerase II 2. Their expression is generally very specific to a certain tissue or in response to external stimuli. MicroRNAs are short, 18 to 25 nucleotides, and generated by enzymatic excision of a larger precursor named primary miRNAs (pri-miRNAs or pri-miRs; Figure 1). A putative regulatory function of pri-miRNAs was not known until the recent discovery by the group of Jean-Philippe Combier in Castanet-Tolosan (France) published in April in Nature journal showing that non-coding pri-miRNAs can encode peptides 3.
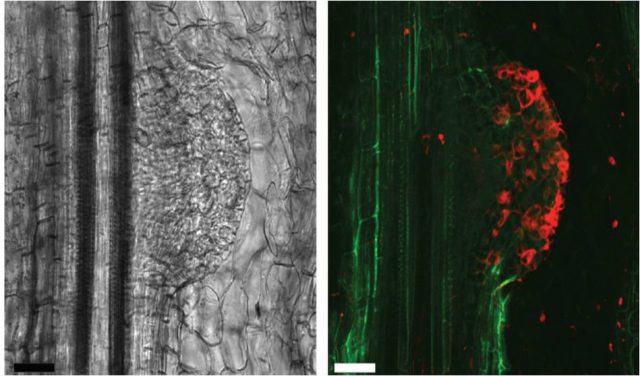
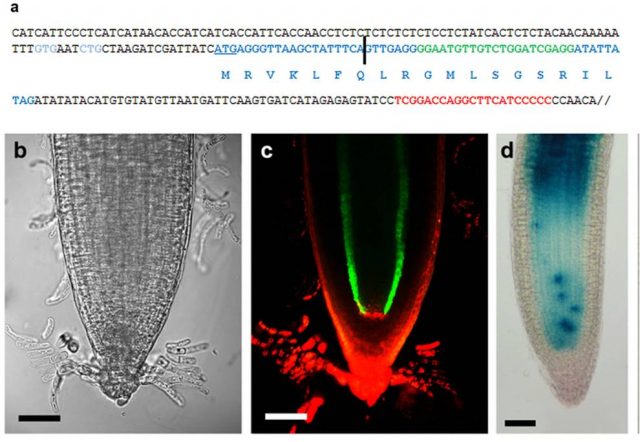
Lauressergues and coauthors started studying the miR171 miRNA family in the legume plant Medicago truncatula because a member of this family miR171b regulates the formation of lateral roots, making it an easy developmental process to be monitored. Thanks to sequence analysis they identified open reading frames (ORFs) in pri-miR171b, synthesized peptides and raised antibodies for these ORFs and demonstrated that a 66-amino acid length peptide, that they named miPEP171b, was truly existing (Figure 2). Moreover, to check whether the existence of miPEPs from pri-miRNAs was a general capacity of this type of non coding RNAs the authors analyzed 50 known sequences of Arabidopsis thaliana and observed the presence of putative ORFs in all of them and experimentally demonstrated the existence of the 18 amino acid length miPEP165a encoded by Arabidopsis pre-miR165a (Figure 2).
To study the regulatory role of miPEPs the authors observed both by treating Medicago truncatula and Arabidopsis roots with their corresponding synthetic peptides or by the overexpression of miPEP175b in M. truncatula roots that miPEPs positively and specifically regulate the abundance of their corresponding miRNA. Moreover, this regulation seems to be at transcriptional level and not by increasing miRNAs stability because the treatment with RNA synthesis inhibitors suppressed the action of miPEPs. Finally, the specific regulation of miRNAs by their corresponding miPEP was further analyzed for other three miPEPs from Arabidopsis and two from M. truncatula.
The discovery of additional miPEPs will not be an easy task since bioinformatic engines often ignore or filter small ORFs because their occurrence by chance alone, increases exponentially as they get shorter 4. Furthermore, since as shown by Laurassergues et al. (2015) these miPEPs can be formed by less than ten amino acids. Overall, miPEPs constitute a novel category of small regulatory peptides in plants, which might be playing a fundamental role for miRNA regulation. It is still to be understood if miRNA transcription activation by miPEPs is constitutive or an inducible process in response to a signal. Future studies will reveal whether miPEPs also exist in animals.
References
- Ariel F, Romero-Barrios N, Jégu T, Benhamed M, Crespi M (2015) Battles and hijacks: noncoding transcription in plants. Trends in plant science. http://dx.doi.org/10.1016/j.tplants.2015.03.003 ↩
- Jones-Rhoades MW, Bartel DB, Bartel B (2006) MicroRNAs and their regulatory roles in plants. Annual Review of Plant Biology 57:19-53. ↩
- Laurassergues D, Couzigou J-M, San Clemente H, Martínez Y, Dunand C, Bécard G, Combier JP (2015) Primary transcripts of microRNAs encode regulatory peptides. Nature 520:90-93 DOI: 10.1038/nature14346 ↩
- Waterhouse PM, Hellens RP (2015) Coding in non-coding RNAs. Nature 520:41-42 ↩
1 comment
[…] Nunca un nombre estuvo peor puesto que el de basura al ADN basura: se confundió el desconocimiento con la inutilidad. Cada vez se conocen más funciones de esta parte del ADN. Los últimos descubrimientos no solo le asignan capacidades […]