Targeting melanoma: dynamic rewiring of signaling pathways contributes to drug resistance in tumors
The scientific community, in their different fields and specialties, has made tremendous progress regarding human health and how we deal with all the maladies affecting mankind. Particularly in cancer, the Pulitzer Prize-winning book entitled “The Emperor of All Maladies” summarizes in a marvelous way this notion. However, I am convinced that our struggle against cancer is far away from coming to an end. As a researcher, I understand cancer as a poker game in which you have a lousy hand and the next opponent´s movement is unknown. However, instead of pessimism I would like to convey to readers that these first lines are a description of reality and optimism towards the tumor chronicity and the improvement of patients’ lifespan.
Cancer is characterized by a genotype and an epi-genotype that explain its aggressiveness and progression. So we focus on it and decipher the molecular codes to unveil the singularities of cancer. However, gladness in cancer research is something ephemeral given that tumor cells quickly manage to evade our acquired knowledge for years and the therapies that once seemed to be successful are not anymore. How tumors evolve and bypass the technology or the drugs we develop is an exciting field in both basic and translational biosciences. This notion is particularly evidenced in those tumor types whose normal tissues came from having an epigenetic and genetic background highly unstable. This is the case of lung cancers or skin malignant melanomas whose original tissues have the highest somatic mutation rates in the body given the tobacco consumption and sun exposition, respectively12. Seeing the huge impact that the environment exerts over these tissues, the epigenetic landscape of these tumors has been investigated. In this line, cell complexity needs also complex mechanisms of transcriptional regulation responding to intracellular stimuli and the external microenvironment. Hence, the collection of all the mechanisms that explain how the cells modify the transcriptome (group of genes expressed under particular circumstances) is what we define as epigenetics (to have a wider view of what epigenetics is, see Javier Carmona´s article “Dog’s DNA methylome uncovers hints on human cancer metastasis” published in this blog).
Particularly, melanoma is one of the solid tumors less epigenetically interrogated. However, huge efforts have been made in the last decade to improve the knowledge we had regarding this aggressive and multi-resistant tumor type. Clinical outcome of the melanoma patients justify this scientific involvement given the remarkable survival difference between localized and metastatic disease (98% and 16-62% 5-year survival, respectively)3. Thus, this setting provided enough stimuli to channel resources into improving the clinical management and diagnosis of melanoma patients.
Epigenetic inactivation of particular tumor suppressor genes by DNA methylation has been extensively implicated in tumorigenesis and progression of a variety of different malignancies. It was the first epigenetic alteration studied in melanoma occurring preferentially in gene promoter regions. Promoters are CG enriched portions of the genes involved in the transcription and susceptible to be silenced by the deposition of methyl groups in the cytosines closed by guanines. First reported genes regulated by DNA methylation in melanoma were PTEN (methylated in ~60% of melanomas45), SYK (methylated in 30% of melanomas6), TNFSF10D and LOX (methylated in 80% and 50% of melanomas7) or COL1A2 (methylated in 80% of melanomas[6]). Even though the first discoveries were made on disrupted genes by DNA hypermethylation, epigenome-wide DNA methylation landscape of melanoma revealed a global wave of hypomethylation affecting intergenic regions, gene bodies and locations away from CpG islands (genome regions with enriched CpG sites). Actually, Guo et al. (2015) found, comparing the epigenomes of primary cutaneous melanoma tumors with different progression states, that the 98% of the disrupted methylated CpG sites were events of loss of methylation whilst only a 2% were events of methylation gain8.
Focusing on the study that concern us today, Dr. Esteller´s team described an epigenetic reactivation of a gene called TBC1D16 involved in the endosomal trafficking (one of the most relevant signaling pathways in melanoma) tightly associated with melanoma progression9 (Figure 1).
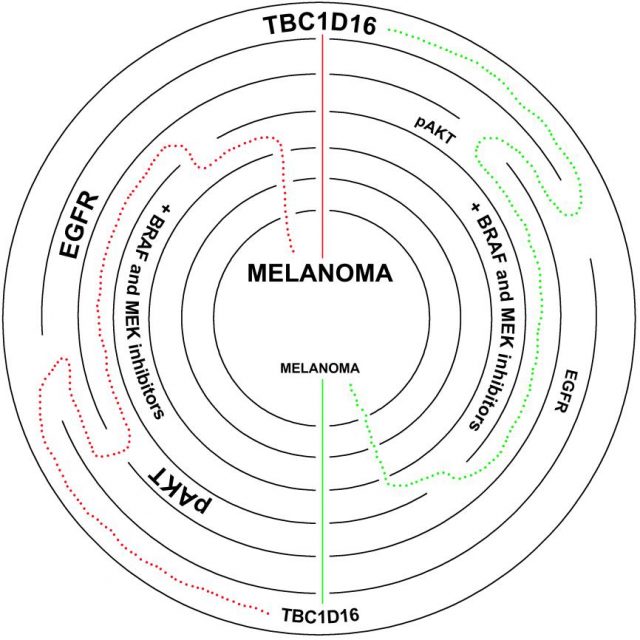
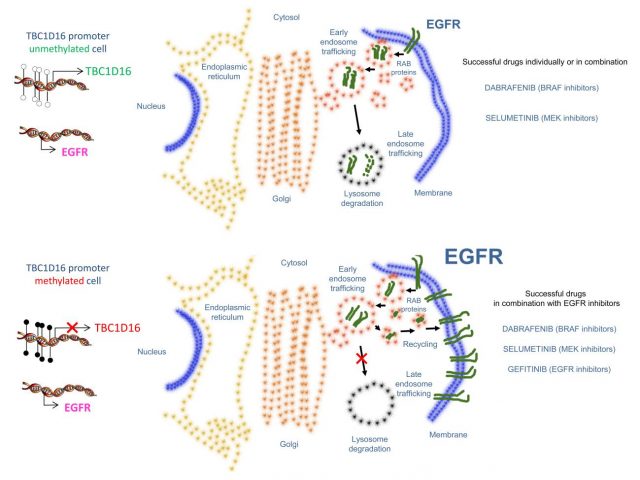
Chronologically, authors reported (i) the reactivation of TBC1D16 is driven by a DNA demethylation event in an inner promoter, affecting to an internal-shorter variant and not to the main-longest isoform pointing up how complex the genome can be, and (ii) the clear association of this reactivation with acquisition of metastatic properties. The authors aim to identify new melanoma biomarkers to improve the clinical outcome of the melanoma patients since the very early stages of the disease. Particularly in melanoma, it is important to know that surgical approach is the best approach we actually have to stop the progression of the disease and only adjuvant therapy with interferon (which has been proven to have high toxicity rates and few benefits) has been approved. Thus, the authors also completed their study looking not only for biomarkers but also for predictive genes of target therapy response. Thus, the researchers finally found a marker that contributes to the clinical stratification of melanoma patients with similar accuracy to other prognosis factors used into the clinics such as Breslow (tumor thickness) or ulceration, but also a useful tool that indicates which patients will completely respond to the ongoing used small drugs Vemurafenib and Dabrafenib. The authors provided experimental data to demonstrate how tumors, depending if they express or not the protein TBC1D16, are more or less addictive to the MAPK/RAS/RAF pathway given the control this protein exerts over the expression of the epidermal growth factor receptor (EGFR) (Figure 2). Worth to mention that EGFR protein is a master regulator of the PI3K/AKT survival signaling which is the contralateral survival pathway of MAPK/RAS/RAF cascade. Thus, this discovery offers new alternative ways to block the acquisition of resistance that the malignant cells manage by “dynamic rewiring” using combinatorial therapies with MAPK/RAS/RAF and EGFR/PI3K/AKT inhibitors.
References
- Govindan R, et al. Genomic Landscape of Non-Small Cell Lung Cancer in Smokers and Never-Smokers. Cell 2012;150:1121–1134. ↩
- Martincorena I, et al. High burden and pervasive positive selection of somatic mutations in normal human skin. Science 2015;348:880-886. ↩
- ACS. American Cancer Society, Cancer Facts and Figures, 2014. (Atlanta: American Cancer Society). ↩
- Mirmohammadsadegh A, et al. Epigenetic silencing of the PTEN gene in melanoma. Cancer Res 2006;66:6546-52. ↩
- Lahtz C, et al. Methylation of PTEN as a prognostic factor in malignant melanoma of the skin. J Invest Dermatol 2010;130:620-2. ↩
- Muthusamy V, et al. Epigenetic silencing of novel tumor suppressors in malignant melanoma. Cancer Res 2006;66:11187-93. ↩
- Liu S, et al. Identification of novel epigenetically modified genes in human melanoma via promoter methylation gene profiling. Pigment Cell Melanoma Res 2008;21:545-58. ↩
- Guo X, et al. In-depth genomic data analyses revealed complex transcriptional and epigenetic dysregulations of BRAF (V600E) in melanoma. Mol Cancer 2015;14:60. ↩
- Vizoso M, et al. (2015) Epigenetic activation of a cryptic TBC1D16 transcript enhances melanoma progression by targeting EGFR. Nat Med. DOI: 10.1038/nm.3863 ↩