Ancient DNA Calling Out for “De-Extinction” — How far can or should we go?

Ever since the 1993 film based on Michael Crichton’s novel Jurassic Park was released, the thought of reviving extinct species using molecular biology techniques has been on the forefront of the collective imaginary. The idea seemed pretty simple: reading the genetic code of fossilized animals would provide the instruction manual to bring them back to life! Skeptics would claim –with all good reason- that it takes more than DNA to make a life. However, intrepid biotechnologists started playing around with the idea. In the end it turns out that things are not as simple –nor as complicated- as they seemed in the beginning. And so, scientists are getting pretty close!
Ancient DNA (aDNA), i.e., genetic material from long-dead biological specimens, has centered the attention of scientists for decades as a way to take a peek into the past. First sequences on ancient DNA were released in the 80’s –the pre-PCR era- using genetic material extracted from museum specimens and thousands of years-old Egyptian mummies 12. These reports raised great enthusiasm and soon many others followed. However, with the advent of the PCR, many published supposedly archaic sequences were found to reflect contamination of DNA from fungi, bacteria or even modern humans that had been in contact with the specimen. These mistakes were not completely meaningless as they led to adopt a series of good laboratory practices to avoid contamination and ensure the authenticity and robustness of the data resulting from studying ancient biological samples 3. DNA degradation over time was another major concern in aDNA sequencing studies. The chemical bonds between nucleotides in the double helix degenerate as a result of UV radiation, temperature, and hydrolysis, and thus DNA exposed to environmental degradation is not likely to survive longer than around 1 million years4. This of course rules out sequencing 65 millions year-old dinosaur’s DNA –that, by the way, has never been found. However, subzero environments offer better conditions for preserving DNA as it slows down its degradation by liquid water or microorganisms. Thus, buried animal remains, especially those found in permafrost layers in Siberia or North America, represent great sources of DNA to recreate life in oblivion.
Ancient DNA Tells Many Stories
The oldest genome that has been sequenced to date belongs to a 700,000-year old horse bone fossil that was recovered from primeval permafrost found in Yukon, in western Canada 5. By comparing its genome with that of modern horses and other related species, scientists tracked the common ancestor of the Equus lineage –horses, donkeys and zebras- to 4 million years back, and exposed the strong influence that human domestication had in shaping their genome. The work broke the record for the oldest complete genome sequenced up to date, which belonged to a 120,000 year-old polar bear specimen. In this case, scientists were able to correlate the divergence between species of brown and polar bear with a period of environmental change during the Pleistocene. Glaciations drove several adaptations to Arctic climates in polar bears –fatty-acid metabolism, hibernation-, whereas brown sister species continued their evolutionary history at lower latitudes 6. Ancient DNA studies have also investigated the causes of mass extinctions of mega-herbivores coinciding with humans entering the scene in the Late Quaternary (50,000 years ago). An extensive study integrating demographic, climatologic, and genomic data from that period reconstructed the influence of climate and humans on the declining populations of woolly rhinoceros, woolly mammoth, horse, caribou, bison and musk ox. The conclusions of the work indicate that the expansion of some human populations and changes in climate influenced the population dynamics of these animals, yet each species responded differently to environmental pressures and no direct cause could be identified 7.
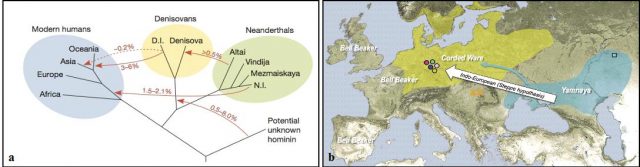
Ancient DNA also tells the story of humankind. Genome-wide analysis of prehistoric human remains has greatly contributed to our understanding of how proto-humans adapted to the different environments they encountered as they spread across the globe. According to the fossil record, first Homo erectus explorers set foot out of Africa 1.8 million years ago and expanded through Eurasia. Subsequently, other groups followed and finally Homo sapiens flooded to the near East some 125,000 years ago. During this time, different human lineages have emerged and gone through population bottlenecks and even extinction along the course of history. However, it was thought that modern human origins were homogeneous and that their –our- evolutionary success was based on the replacement of pre-settled hominin populations such as Neanderthals. In 2014, two ground-breaking studies proved that in fact modern humans have a more complex genomic make up. As they migrated out of Africa, Homo sapiens interbred not only with Neanderthals —1.5 to 2.1 % of the DNA of modern populations is Neanderthal in origin-, but also with Denisovans –that left their genetic imprint in Australasian populations-, and even some other as-yet-unknown archaic hominid populations that diverged early from the main human lineage 8 9 (Figure 1). Human evolution is not as homogenous as once thought. In fact, cross-breeding between different hominid species might have provided modern humans with improved abilities to cope with cold climates or solar radiation. Similar studies have shown that genetic traits fixed in modern populations can help to solve the conundrum of migratory pulses from Siberia which gave rise to modern Native-Americans some 23,000-13,000 years ago 10; or shed light on the origins of Indo-European languages in ancient nomadic cultures that travelled across Europe in the early Bronze age (3,000BC to 1,000BC) 11 (Figure 1).
Resurrecting Old Relatives
Besides providing information on how ancient living communities evolved and interacted, studying aDNA has fostered a new area of research dubbed “De-extinction” that focuses in bringing vanished species back to life. The premise behind it is that the recovery of DNA from an extinct species would provide the starting point for its regeneration in a laboratory. There are different technical approaches that could theoretically be used to this end. The first approach, somatic nuclear cell transfer (SNCT), was used to clone Dolly the sheep. It consists on inserting the genetic material of the individual to be cloned inside an enucleated egg from a living relative, implanting it in a surrogate host and bring pregnancy to terminus. This procedure was successfully applied to de-extinct the Iberian wild goat (Capra pyrenaica pyrenaica; Figure 2) in 2,000, a few years after the last specimen died in the Spanish Pyrenees. Conservational biologists managed to obtain fibroblasts from the last living animal, subcultured them in vitro and injected their nucleus into enucleated domestic goat oocytes. One of the implanted embryos reached the end of the gestation and was brought to life but it died within a few minutes due to a defect in its lungs 12. Nevertheless, the project is still active and aims to restore a population of Iberian wild goats in its original environment.

Other projects currently ongoing aim to resurrect the Tasmanian tiger, the Australian gastric-brooding frog, or the brown-and-white-striped quagga, among others (Figure 2). However, since many of these animals have been extinct for long periods of time, no viable cells remain to carry out SNCT protocols. Hence, scientists have thought of an alternative approach to overcome this pitfall. In theory, by editing the endogenous genome of a stem cell from the closest living relative of an extinct species, it would be possible to reprogram it to give rise to its ancestor. This is possible thanks to a recently discovered molecular biology cut-and-paste tool, CRISPR/Cas9, which can target specific regions in the genome and modify them at will. Thus, identifying the base genome of a living descendant, and spotting the exact genomic changes to be introduced are the two ingredients to kick-off a de-extinction project. In the last few years, the project of resurrecting the woolly mammoth has focused on these two goals.
The complete nuclear genome sequence of the extinct woolly mammoth was deciphered in 2,008 at a resolution comparable to that of modern genomic studies13. Thanks to next-generation sequencing protocols, authors were able to make the most of the DNA recovered from incredibly well-preserved mammoth bones and mummies found in the permafrost of the Arctic tundra, and succeeded in assembling the first complete genome of a pre-historic animal. The results of this study showed that the woolly mammoth and the modern African elephant diverged 6 million years ago, but share 99% of their DNA. Scientists also mapped a handful of non-synonymous changes in protein-coding genes unique to the woolly mammoth that improved its metabolic and morphological adaptation to glacial temperatures –woolly coats, subcutaneous body-fat, or hemoglobin for oxygen transport in subzero environments. Going forward, Dr. George Church and his team at Harvard Medical School are currently using CRISPR technology to splice those functional changes into the genome of modern African elephants hoping to generate hybrids displaying the same adaptations to cool temperatures that mammoths once had. However, as Dr. Church pointed out, in the hypothetical scenario that this whole process is technically feasible, the result will never be an exact mammoth duplicate. Single-Nucleotide Polymorphisms (SNPs) in other parts of the genome, or epigenetic factors responding to environmental stimuli and biological interactions, play a regulatory role that remains unknown and will not be reproduced on de-extinct animals.
Should We Plunge Into a Re-Evolution?
Using modern genomics, several species of thousands of year-old are “on their way to resurrection” in different laboratories around the world. It seems that the technical means to revive extinct species are around the corner, yet there are still profound questions that need to be addressed. These come in two flavors: practical –which will be eventually overcome-, and philosophical.
Scientists in favor of de-extinction assert the moral imperative of bringing back species that have been erased by human action in the last 10,000 years, in addition to the potential ecological benefits of bringing back these animals. From a technical perspective, it might be easier to resurrect animals that have been gone for few decades –and for which there are frozen viable cells and tissues that can be manipulated- than to bring back pre-historic species. However, certain unpredictable issues include what influence would these animals have on current ecosystems; how they would compete with co-existing species some of which might even be presently endangered; and importantly, what would be the consequences of resurrecting an almost-equal –but not exact- copy of a once-living animal. Other voices however argue that the technology of de-extinction should be instead used to help preserving endangered species that have not yet disappeared (Figure 3).

Conservational biologist Beth Shapiro proposes using genome-editing technology –CRISPR- to modify the genome of declining Asian elephant or white rhinoceros populations in a way that allows them adapting to cooler environments, thereby expanding the habitats on which they can live and thus increasing their chances of survival. In Hawaii, Dr. Ruth Gates and her group have been studying the decline of coral reef communities over the past years. In addition to artificially growing reefs under controlled conditions, her team is identifying genetic traits that could be spliced into endangered species to make them resilient to heat and acid environments resulting from global warming.
Thus, if the technology reaches a point that allows doing it, should this be carried out? Scientists do not seem to reach a consensus. Ethical aspects and real motivations behind such actions are not trivial and need to be weighed. Moreover, adapting the legislation to regulate genetic engineering practices is sorely needed. Turning genetic blueprints into living organismsfaces many unresolved questions besides the technological limitations and further research and debate will be needed to evaluate all possible angles and outcomes.
Truth is stranger than fiction. With the fast-pace at which technology advances, we may end up facing de-extinction in real-life as was anticipated on movies. In the meantime, sit and enjoy.
References
- Higuchi R et al. DNA sequences from the quagga, an extinct member of the horse family. Nature (1984) 312, 282 – 284. ↩
- Pääbo S. Molecular cloning of Ancient Egyptian mummy DNA. Nature (1985) 314, 644 – 645. ↩
- Cooper A and Poinar HN. Ancient DNA: Do It Right or Not at All. Science (2000) 289 no. 5482 p. 1139. ↩
- Allentoft ME et al. The half-life of DNA in bone: measuring decay kinetics in 158 dated fossils. Proceedings of the Royal Society B. (2012) DOI: 10.1098/rspb.2012.1745. ↩
- Orlando L et al. Recalibrating Equus evolution using the genome sequence of an early Middle Pleistocene horse. Nature (2013) 499, 74–78. ↩
- Miller W, et al. Polar and brown bear genomes reveal ancient admixture and demographic footprints of past climate change. Proc Natl Acad Sci U S A. (2012) 109(36):E2382-90. ↩
- Lorenzen ED et al. Species-specific responses of Late Quaternary megafauna to climate and humans. Nature (2011) 479, 359–364. ↩
- Meyer M et al. A mitochondrial genome sequence of a hominin from Sima de los Huesos. Nature (2014) 505, 403–406. ↩
- Prüfer K, et al. The complete genome sequence of a Neanderthal from the Altai Mountains. Nature (2014) 505, 43–49. ↩
- Skoglund P et al. Genetic evidence for two founding populations of the Americas. Nature (2015) doi:10.1038/nature14895. ↩
- Haak W, et al. Massive migration from the steppe was a source for Indo-European languages in Europe. Nature (2015) 522, 207–211. ↩
- Folch J et al. First birth of an animal from an extinct subspecies (Capra pyrenaica pyrenaica) by cloning. Theriogenology (2009) 71(6):1026-34. ↩
- Miller W et al. Sequencing the nuclear genome of the extinct woolly mammoth. Nature (2008) 456, 387-390. ↩
3 comments
[…] Las posibilidades técnicas para resucitar especies animales extintas están a la vuelta de la esquina, ¿deberíamos hacerlo? F. Javier Carmona en Ancient DNA Calling Out for “De-Extinction” — How far can or should we go? […]
Interesante artículo. Muy bueno el comentario del oso.
Pero…, tóesto lo has escrito tú???
Amazingly interesting, please more.
Final thought left rather disquieting though!
Ain’t we too much futurelooking whilst our present is derailing, thwarting?
Shoudn’t we shift into a kind of sustainable degrowth?
FAO works as Save and Grow lead us how.