Graphene nanopore DNA sequencing
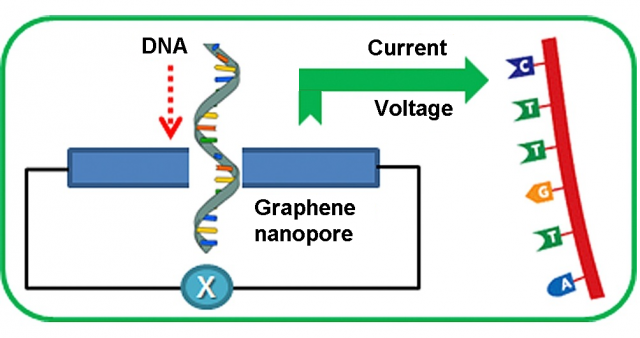
Nanopore DNA sequencing was one the ten scientific breakthroughs of 2016 highlighted by Science magazine. In principle, graphene is the perfect pore material for DNA sequencing 1. Its monoatomic thickness of 0.35 nm is similar to the DNA base spacing and graphene nanopores can be fabricated with a diameter of only 1.0 nm, about the size of a DNA molecule. Even if currently there are some drawbacks to be addressed, it is one of the most promising and revolutionary nanopore DNA sequencing technologies.
The sequencing of a DNA molecule consist of reading in proper order its sequence of letters, the four nucleobases cytosine (C), guanine (G), adenine (A), and thymine (T). DNA sequencing methods have been classified in four generations: first, Sanger sequencing, second, amplification-based massively parallel sequencing, third, single-molecule sequencing, and fourth, nanopore sequencing. The last promises inexpensive, reliable, and high-throughput sequencing.
Nanopore sequencing was first demonstrated in 1996 by Deamer, Branton and their colleagues 2. They used a α-hemolysin biological channel/pore; a distinct current signal was observed depending on the passage of purines (A and G) or pyrimidines (C and T). Further advances in α-HL-based technology result in single DNA sequencing in 2012, the year the company Oxford Nanopore Technologies is born. As an alternative, solid-state nanopores were first introduced in 2003 for DNA sequencing 3. They were made of silicon nitride, but today other materials are also used, like silicon oxide, aluminum oxide, polymer membranes, glass, hafnium oxide, gold, etc. However, up to now, DNA sequencing with a single base resolution has not been achieved by using of solid-state nanopores.
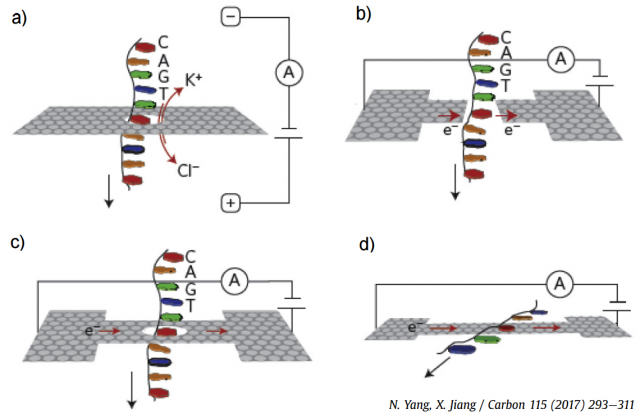
Graphene nanopores, nanogaps and nanoribbons have been employed for DNA sequencing, as illustrated in the four plots in Figure 2. The most efficient approach is the measurement of the ionic blockade current produced when a single-stranded DNA translocates through a graphene nanopore (Fig. 2(a)), since the different bases yield distinctive fluctuations. Figure 2(b) shows a theoretical proposal, since no real device is currently available, the modulation of the fluctuations in a transverse tunneling current when DNA passes through graphene nanopore or nanogap. The in-plane current through a graphene nanoribbon also changes when a DNA molecule traverses a nanopore in its middle, as shown in Fig. 2(c). Finally, graphene-DNA interactions, as shown in Fig. 2(d), can also be exploited in nanoribbons without pores. There are other alternative approaches that have been proposed and tested experimentally, but they are less developed.
Obviously, graphene nanopore sequencing also has drawbacks. The biggest one is that DNA molecules stick strongly to graphene surface and nanopores. The only solution is the passivation of the surface of the graphene. Additionally, the low-frequency noise in graphene nanopores is typically two orders of magnitude higher than for silicon nitride nanopores. This is due to the low mechanical stability of a graphene membrane as a result of its atomic thickness resulting in elastic corrugation and the appearance of defects. Finally, do not forget that current fabrication techniques result in a irreproducible and irregular geometry for graphene nanopores.
In summary, graphene nanopore sequencers are actually in the R+D phase, so further research is required. Nothing prevents us from dreaming a future sequencer consisting of a detector chip, an array of graphene nanopores, an integrated microfluidic system, electronic probes, and a portable system. Will future CSI investigators have a graphene DNA sequencer incorporated into its own mobile phone?
References
- N. Yang, X. Jiang, “Nanocarbons for DNA sequencing: A review,” Carbon 115: 293–311 (May 2017), doi: 10.1016/j.carbon.2017.01.012. ↩
- J. J. Kasianowicz, E. Brandin, …, D. W. Deamer, “Characterization of individual polynucleotide molecules using a membrane channel,” PNAS 93: 13770–13773 (1996), PubMed ID: 8943010. ↩
- J. Li, M. Gershow, …, J.A. Golovchenko, “DNA molecules and configurations in a solid-State nanopore microscope,” Nature Materials 2: 611–615 (2003), doi: 10.1038/nmat965. ↩
1 comment
[…] La secuenciación del ADN usando nanoporos es uno de los hitos científicos de 2016. El grafeno puede ser el material ideal para construir los nanoporos necesarios. Francisco R. Villatoro en Graphene nanopore DNA sequencing […]