Epigenetic alterations in Alzheimer’s disease, nature vs. nurture on the path to dementia
Nowadays there are still people that believe in destiny. However the scientific community more and more is bringing light to that subject showing that although the genetic material could program ourselves to suffer some pathologies, the day-to-day experiences are the ones that lead us towards a healthy or pathological aging. And how is that? Our DNA suffers mutations that alter the gene expression and protein function but the real modulators of the proteome of our cells are epigenetic mechanisms. Epigenetic was a term coined by Chris Weddington in 1942 referring the way genes interact with their milieu in order to generate a phenotype. Today is one the hottest topics in biomedical research. Epigenetic processes are major effectors of gene expression regulation and they involve complex series of mechanisms like DNA alterations (i.e. methylation), histone modifications (i.e. phosphorylation, acetylation) and the actions of non-coding RNA (i.e. microRNA, lncRNA, piRNA). Deregulated epigenetic patterns are being associated to many human diseases, especially cancer but recently is more evident their role in brain disease such as depression or neurodegeneration.

Alzheimer’s disease (AD) is an age-associated neurodegenerative disorder, characterized by cognitive decline and memory loss. AD is the leading cause of age-related dementia. Present demographic trends, especially with respect to life expectancy, call for the rapid development of more effective preventative and therapeutic approaches in order to reduce suffering and costs associated with the disease; the World Health Organization (WHO) estimates that 100 million people will suffer from AD by 2050. There are two classes of that neurodegenerative disease, the familiar form (FAD) and the sporadic AD (SAD). The FAD is due to genetic mutations in some genes like ApoE, APP or PSEN1/2. However, this form of the disease represents only around 5% of the total. Familial AD is an heritable disease, but SAD is not. Although, we often found several AD cases in the same families which are not due to genetic mutations (important to mention is that FAD form is very aggressive and normally is present between 50 and 65 years of age). One possible explanation of why this occur could rely on environmental factors that would induce epigenetic alterations programming the subject to the disease (although it could also be explained by the fact that we often diagnose as AD any type of dementia since there are no methods for detection of the disease before death; but that is a story for another post).
Therefore understanding the possible epigenetic alterations through lifespan is one of the biggest challenges for the scientific community on the field of AD. Here is where Esteller’s group1[PDF] brought some light to the messy neuroscience. Sanchez-Mut et al. focuses into analyze epigenetic alterations in specific mouse brain areas by using two of the most studied animal models of the disease (APP/PSEN1 model and 3xTg-AD) and comparing the results obtained with human brain samples from postmortem tissue from AD patients at advanced stages of the disease. Is important to mention that brain studies imply a highly complicated heterogeneity due to the spatial specificity of the organ. However, to overcome that issue the authors used in an unprecedented way, a high brain region analysis, punching the 12 most relevant brain areas of the mice (among them frontal cortex and hippocampal regions, key areas in AD pathophysiology), and through the sequenciation of the epigenome via genome wide bisulphite sequencing, they analyzed the methylomic profiling of 384 mouse genes in each of those 12 brain areas.
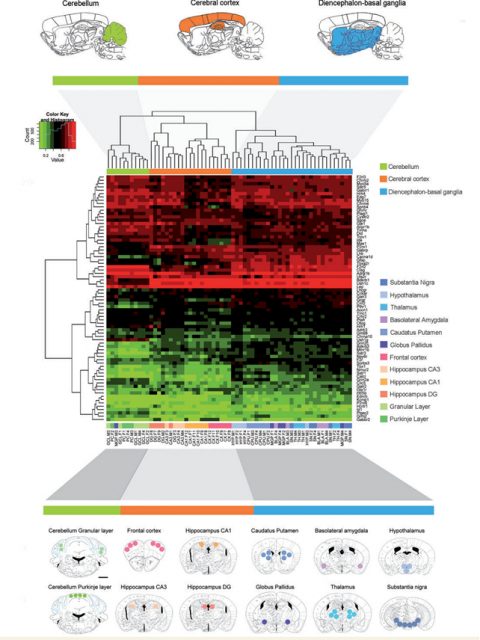
By hybridizing the DNA obtained from the punches of each of the brain areas analyzed to a custom microarray containing the 5’-end regulatory regions of preselected genes, the authors were able to identify an important list of genes, specially related to processes like neuroplasticity, cognition and related to mental disease. These data were validated by pyrosequencing analysis and the array data were correlated to RNA expression profiles. Esteller’s team found a high correlation between DNA methylomic profiling and RNA expression patterns, demonstrating the biological significance of their findings.
As mentioned before, different brain areas show different properties due to different cell populations and function. Thanks to the approach described above and developed in wild-type animals, the team identified subsets of genes that were differently regulated accordingly to the spatial localization. From those, a subset of genes were predominantly altered in cerebral cortex regions. Cerebral cortex is a brain area highly involved in AD (related to memory formation and spatial navigation, both cognitive aspects are severely affected in dementia). From the list of genes differentially methylated that the authors described in their analysis, 52 were different between cerebral cortex and the rest of the brain. To determine whether any of the genes of the frontal cortex subset were affected in AD, the authors hybridized frontal cortex samples from 2 well studied mice models of the disease. Interestingly, 4 genes (Tbxa2r, F2rl2, Sorbs3 and Spnb4) were hypermethylated (and thus down-regulated) in the frontal cortex of both models, highlighting their importance on the disease pathogenesis in that specific brain region.
The final goal of biomedical research is always trying to find cures for human diseases, and thanks to the experience of Esteller’s lab with human samples, the authors pursued the translation of their findings to AD patients. To achieve that, they analyzed those 4 genes altered in frontal cortex in 2 of the better studied AD mouse models in a collection of 20 samples extracted from frontal cortex of AD patients on Braak V-VI stage. Braak staging is a method to classify the severity of AD (also used for Parkinson). That classification starts at level I and finishes at stage VI, being early and late stages of the disease respectively. Is mainly in stages V and VI when the frontal cortex of the patient is severely affected. The findings of Dr. Jose V Sanchez-Mut and colleagues conclusively demonstrated that the 4 genes downregulated in mice were severely affected in humans as well. These results were confirmed by a decrease of the RNA levels of those genes and a reduction of the protein concentration.
But what are these genes? What is their role in the cell homeostasis? The 4 genes described in mice and humans are related to neuronal function and its role in neuronal communication. TBXA2R is a regulator of CREB, a very important molecule on neuron activity. Also plays a role, together with F2RL2, mediating neuronal toxicity induced by NMDA, a process associated with the neurodegeneration observed in AD. SORBS3 is associated with synapsis regulation, the basic unity on neuronal communication and SPTBN4 is essential also for synaptic integration and communication. Thus, all 4 genes are involved with one of the most affected units in AD, the neuronal synapses and the integrity of the neurons. The findings published by Sanchez-Mut and col., open a new possibility for finding AD treatments, suggesting 4 new targets for pharmacological interventions.
AD is not anymore a disease considered only present in old people, because more and more, the data indicates that it starts around 20-30 years before the onset of the pathology is evident. Thus, is crucial to understand when and why those 4 genes, among others, start to be deregulated and if blocking the hypermethylation of those we can delay or prevent the development of the disease. Our chess game againsts AD will be long and savage and the scientific community is doing just the first movements, but thanks to papers like the one published by Esteller’s team, checkmate to AD is closer now than before.