How drug discovery works: finding new strategies against Kaposi’s sarcoma
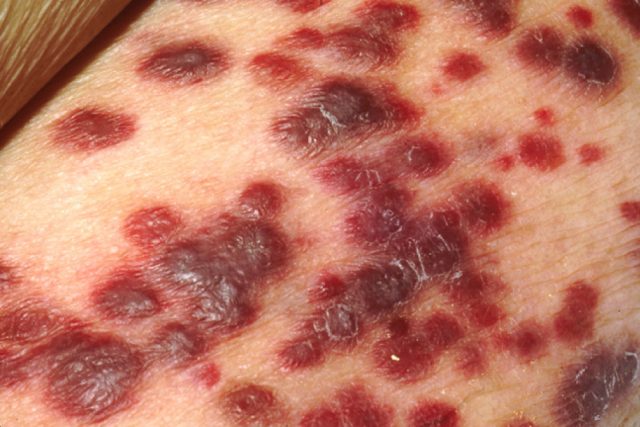
15% of human cancers worldwide may be attributed to viruses,1 with both DNA and RNA viruses being capable of causing the disease. Epstein-Barr virus, human papilloma virus, hepatitis B virus, and human herpes virus-8 are the four DNA viruses capable of causing the development of human cancer, whereas human T lymphotrophic virus type 1 and hepatitis C virus are the two RNA viruses that contribute to it.2 Kaposi’s sarcoma is an example of cancer caused by a virus: the Kaposi’s sarcoma-associated herpesvirus (KSHV, also known as human herpes virus-8) that belongs to the family of human herpesviruses, which are responsible for a range of debilitating acute and recurrent diseases, including a number of cancers.3 4 5 KSHV affects mainly immunocompromised patients such as people undergoing transplants or HIV patients. It was relatively recently (1994) that KSHV was identified in biopsies from tumours of a patient with Kaposi sarcoma,6 a relatively rare disease prior to the AIDS epidemic. It causes patches of abnormal tissue to grow under the skin and in the mouth and is a cancer of urgent and unmet medical need, particularly in the developing world.
Recently, Sophie Schumann and her colleagues have published a study7 on KSHV that might have many implications in the future, not only for the treatment of Kaposi’s sarcoma but for the inhibition of a number of viruses. To date, the treatments for herpesviruses had been limited to targeting the DNA polymerases (i.e. enzymes responsible for the synthesis of the virus’s DNA). But with emerging viral resistance and little efficacy against the cancer-causing herpesviruses, new therapeutic approaches are needed 8. So Schumann’s group focused their efforts in finding different strategies to attack herpesviruses in a different way.
Like all herpesviruses, KSHV has two possible forms of infection: latency (a dormancy phase in which, after initial infection, proliferation of virus stops but the viral genome is not eradicated and the virus can reactivate and replicate itself within the host, without cells being infected from the outside) and lytic replication (a destructive process in which the virus injects its own genetic material into a host cell and then hijacks the cell’s replication and translation mechanisms, using them for its own benefit to make more viruses; once a number of them have accumulated, the cell bursts due to high internal osmotic pressure and this releases the new viruses ready to go on to infect the surrounding cells). 9 10
Schumann and her colleagues thought of the lytic replication as a good therapeutic target for the inhibition of the KSHV. One of the key factors of the lytic cycle is ORF57, a well-known protein present in KSHV that is essential for viral replication.11 12 Through a series of complex biochemical processes, the interaction between ORF57 and the human protein complex hTREX13 produces viral ribonucleoprotein particles (vRNPs, formed by viral proteins and genetic material) that are essential for the lytic replication of the virus. 14 15 16
It is not difficult to imagine that, at least in theory, by disrupting the interactions between ORF-57 and hTREX, the lytic cycle of the virus could be halted and therefore the infection stopped. However, it is not that straightforward: hTREX is a multiprotein complex formed by several components. Being the well-known protein UAP56 one of them, the authors decided to focus their efforts on this part of hTREX.
ATP now comes into play: As many of us may remember from secondary school, ATP (adenosine triphosphate) is the “energy currency” of biochemical systems. When ATP breaks, it yields ADP (adenosine diphosphate), one phosphate group, and energy that can be subsequently used in biochemical processes.
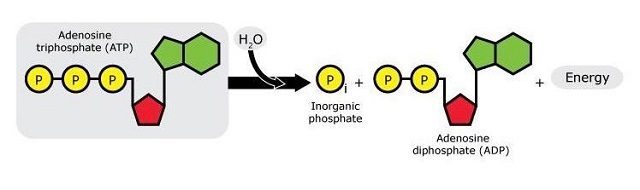
It was previously known that the activity of UAP56 is ATP-dependent.17 This gave the researchers an idea: if they could show that the formation of vNRPs was ATP-dependent too, it might be because it depends directly on the activity of UAP56. Indeed, they were able to show that the hydrolysis of ATP is essential for the ORF57–hTREX interaction: first, they found that ATP binds to hTREX through the UAP56 bit and that, once bound, ATP is hydrolysed to give ADP. The hydrolysis of ATP not only releases energy but also initiates a change of shape in hTREX that allows ORF57 to fit it through the Aly bit (another component of hTREX), like a piece in a jigsaw puzzle. Once ORF57 and hTREX are bound to each other, a series of biochemical processes is initiated, with the synthesis of vRNPs as a result. Inhibiting this activity would therefore prevent the change of shape of the hTREX complex and stop its interaction with the viral protein.
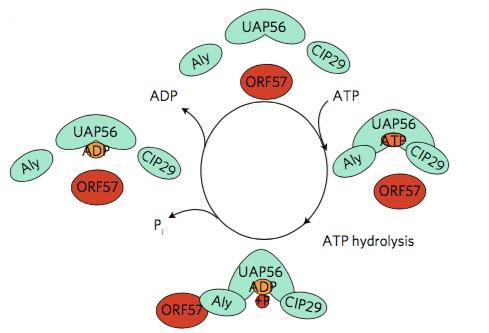
Schumann and her group thought that “tricking” UAP56 into interacting with an inhibitor that was able to displace the ATP molecules, could selectively stop the virus lytic cycle. So the second stage of this study was the in silico high-throughput screening of a number of small molecules. This process consists in the automated testing of a large number (generally several thousands, or even hundreds of thousands) of chemical compounds against the target protein. In this study, a few molecules showed their ability to be ATP-competitive, by occupying the UAP56 ATP-binding pocket (i.e., the area of UAP56 with which ATP molecules interact) by “tricking” UAP56 into making it “believe” they were ATP. A chemical compound called CCT018159 was identified as the most potent, with a reduction of the UAP56 activity of up to 70% depending on its concentration.
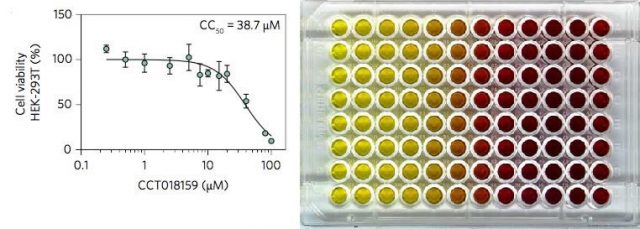
Later, the effect of CCT018159 on vRNP formation was assessed against HEK293T cells (a type of human kidney cells, widely used in routine biology studies) previously infected with KSHV. The cells were incubated in the presence of a series of concentrations of CCT018159 and the results were measured via several methods. The experiments not only showed specific UAP56-inhibition and disruption of vRNP formation, but also that CCT018159 showed minimal toxicity to the cells. The results demonstrated that CCT018159 does not affect other functions of the hTREX protein complex -we should not forget that hTREX is present in healthy cells where it performs vital functions, apart from playing a role in the viral replication when cells are infected. Together, the data suggest that CCT018159 prevents vRNP formation by inhibiting the interaction between UAP56 and ATP whilst not affecting cellular processes, and that cytotoxicity only occurs at higher concentrations than those needed for the inhibition of viral growth. In summary, CCT018159 showed itself able to stop the viruses reproducing, at concentrations that are not toxic to human cells.
Then, the effect of CCT018159 on other herpesviruses was studied. This was justified by all herpesviruses having an ORF57 homologue (a protein with the same activity and very similar structure) that interacts with hTREX for vRNP assembly. Indeed, the replication of HSV-1 (herpes simplex virus type 1, the agent causing cold sores) and HCMV (human cytomegalovirus, associated with infections of the salivary glands in immunocompromised patients and, eventually, mucoepidermoid carcinoma and possibly prostate cancer) was shown to be inhibited by CCT018159 in a dose-dependent manner.
Together, these results indicate that CCT018159 is an inhibitor with possible pan-herpesvirus activity. So far, it cannot be considered a drug, but it is a very good starting point for the development of enhanced, more potent antivirals based on its structure that selectively inhibit the function of UAP56. Now it is on the way to being optimised in order to improve its effectiveness and to reduce the potential for side effects, with the aim of developing a drug that could be used therapeutically in humans, not only as treatment for Kaposi’s sarcoma but for a number of diseases caused by viruses of the same family, including chicken pox and herpes.
References:
1 zur Hausen, H. “Viruses in human cancers”. Science. 254(5035): 1167 (1991). DOI: 10.1126/science.1659743
2 Liao, J. B. “Viruses and human cancer”. Yale Journal of Biology and Medicine 79(3-4): 115 (2006).
3 Gilden, D. H., Mahalingam, R., Cohrs, R. J. & Tyler, K. L. “Herpesvirus infections of the nervous system” Nature Clinical Practice Neurology. 3: 82 (2007). DOI:10.1038/ncpneuro0401
4 Owen, C. B.; Hughes, D. J.; Vaquero-Pérez, B.; Berndt, A.; Schumann, S.; Jackson, B.; Whitehouse, A. “Utilising proteomic approaches to understand oncogenic human herpesviruses” (Review). Molecular and Clinical Oncology 2: 891 (2014). DOI: 10.3892/mco.2014.341
5 Quadrelli, C.; Barozzi, P.; Riva, G.; Vallerini, D.; Zanetti, E.; Potenza, L.; Forghieri, F.; Luppi, M. “β-HHVs and HHV-8 in lymphoproliferative disorders”. Mediterranean Journal of Haematology and Infectious Diseases. 3: e2011043 (2011).
6 Chang, Y.; Cesarman, E.; Pessin, M. S.; Lee, F.; Culpepper, J.; Knowles, D. M.; Moore, P. S. “Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi’s sarcoma”. Science6;266(5192):1865 (1994). DOI: 10.1126/science.7997879
7 Schumann, S.; Jackson, B. R.; Yule, I.; Whitehead, S. K.; Revill, C.; Foster, R.; Whitehouse, A. (2016). “Targeting the ATP-dependent formation of herpesvirus ribonucleoprotein particle assembly as an antiviral approach”. Nature Microbiology, 2: 16201. DOI: doi:10.1038/nmicrobiol.2016.201
8 Bacon, T. H.; Levin, M. J.; Leary, J. J.; Sarisky, R. T.; Sutton, D. “Herpes simplex virus resistance to acyclovir and penciclovir after two decades of antiviral therapy”. Clinical Microbiology Reviews 16: 114 (2003).
9 Jackson, B. R.; Noerenberg, M.; Whitehouse, A. “A novel mechanism inducing genome instability in Kaposi’s sarcoma-associated herpesvirus infected cells”. PLoS Pathogens 10: e1004098 (2014). DOI: 10.1371/journal.ppat.1004098
10 Mesri, E. A.; Cesarman, E.; Boshoff, C. “Kaposi’s sarcoma and its associated herpesvirus”. Nature Reviews Cancer 10: 707 (2010). DOI :10.1038/nrc2888
11 Boyne, J. R.; Whitehouse, A. “γ-2 Herpes virus post-transcriptional gene regulation”. Clinical Microbiology and Infection 12: 110 (2006).
12 Majerciak, V.; Zheng, Z. “KSHV ORF57, a Protein of Many Faces”. Viruses 7(2): 604 (2015). doi: 10.3390/v7020604
13 Schumann, S.; Jackson, B.; Baquero-Pérez, B.; Whitehouse, A. “Kaposi’s sarcoma-associated herpesvirus ORF57 protein: exploiting all stages of viral mRNA processing”. Viruses 5: 1901 (2013).
14 Boyne, J. R.; Jackson, B. R.; Taylor, A.; Macnab, S. A.; Whitehouse, A; “Kaposi’s sarcoma-associated herpesvirus ORF57 protein interacts with PYM to enhance translation of viral intronless mRNAs”. EMBO Journal. 29: 1851 (2010). DOI: 10.1038/emboj.2010.77.
15Jackson, B. R.; Boyne, J. R.; Noeremberg, M.; Taylor, A.; Hautbergue, G. M.; Walsh, M. J.; Wheat, R.; Blackbourn, D. J.; Wilson, S. A.; Whitehouse, A. “An interaction between KSHV ORF57 and UIF provides mRNA-adaptor redundancy in herpesvirus intronless mRNA export”. PLoS Pathogens 7: e1002138 (2011).
16 Boyne, J. R.; Colgan, K. J.; Whitehouse, A. “Recruitment of the complete hTREX complex is required for Kaposi’s sarcoma-associated herpesvirus intronless mRNA nuclear export and virus replication”. PLoS Pathogens 4: e1000194 (2008)
17 Jankowsky, E. “RNA helicases at work: binding and rearranging”. Trends in Biochemical Sciences 36(1): 19 (2011). DOI: 10.1016/j.tibs.2010.07.008