Throwing light on microbial dark matter
Throwing light on microbial dark matter
microbial dark matter
Author: Ramón Muñoz-Chápuli has been Professor of Animal Biology in the University of Málaga until his retirement. He has investigated for forty years in the fields of developmental biology and animal evolution.
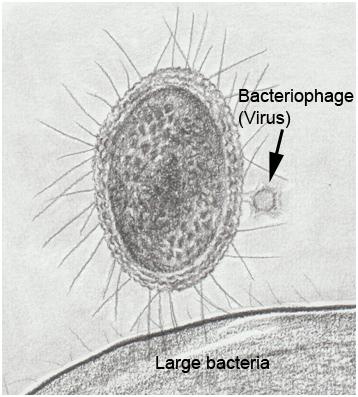
In recent years, single cell genomics and metagenomic studies of environmental samples have enabled insights into “microbial dark matter”, the vast majority of microorganisms (basically Bacteria and Archaea) that cannot be cultured in the laboratory. A significant fraction of microbial dark matter is represented by patescibacteria or CPR (Candidate Phyla Radiation), a phylogenetically primitive group of bacteria with unique characteristics, for example their small size (200-300 nm) and streamlined genome (around 1 Mb, a fifth of the E. coli genome size) (Figure 1). This genome lacks many genes essential for other bacteria, for example those of the Krebs cycle, respiratory chain, and major biosynthetic pathways (amino acids, nucleotides, fatty acids) 1. These features (very small volume, reduced genome size, loss of genes of primary metabolism) also characterize other types of bacteria that live inside host cells, such as Wolbachia, an endosymbiont found in many arthropods, and Mycoplasma, cellular parasites that infect a wide range of organisms and are among the smallest self-replicating organisms. However, patescibacteria do not reside inside other cells; they must be obligate ectosymbionts to obtain nutrients from other microorganisms. Until now, there has been no way to manipulate the genome of any patescibacteria to study their physiological processes.
Cultivating some microbial dark matter
Recently, it has become possible to cultivate some strains of Saccharibacteria, a group of CPR found in terrestrial and marine environments, as well as in the human oral microbiome 2. These strains are dependent on their association with other bacteria. The ability to cultivate them has allowed a group of microbiologists from the University of Washington in Seattle, leaded by Joseph Mougous, to manipulate the genome of a Saccharibacteria species for the first time, specifically Southlakia epibionticum, which lives in association with Actinomyces israelii, the agent of human actinomycosis. This report has been recently published in Cell 3.
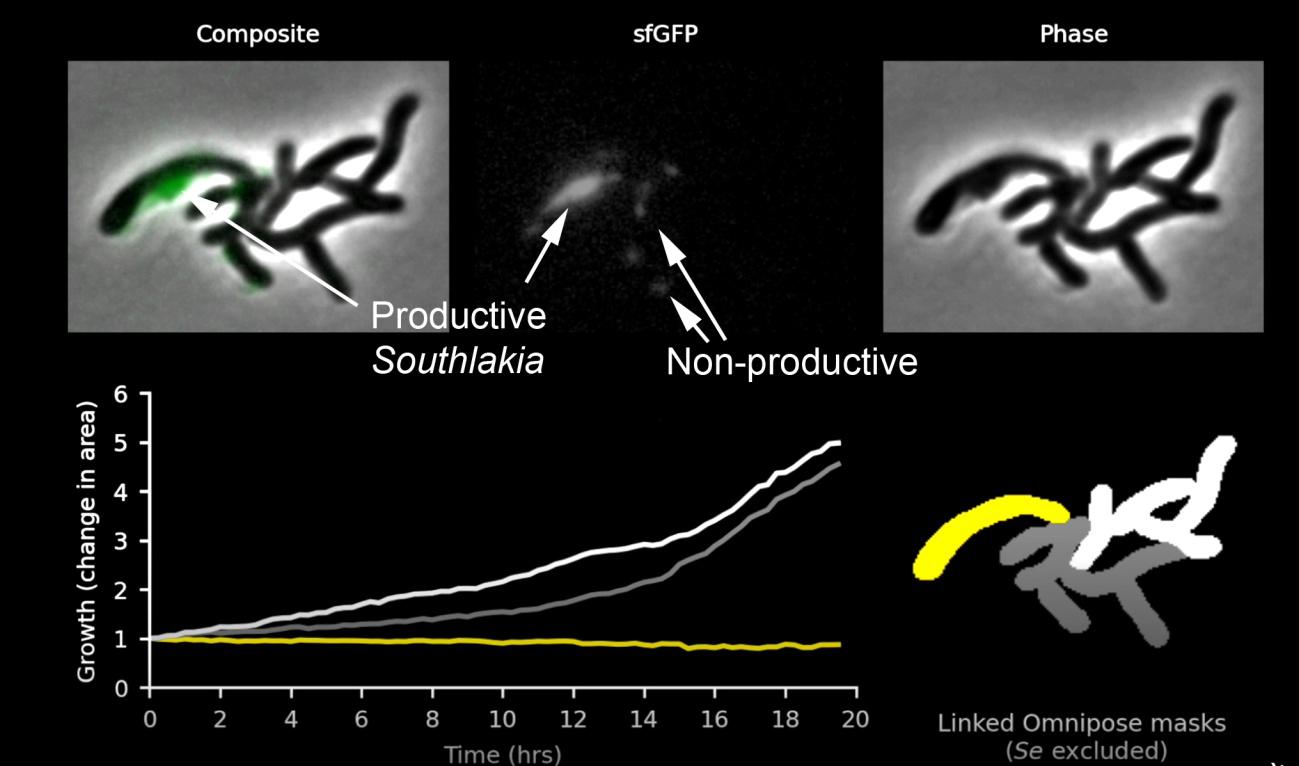
It was already known that patescibacteria were capable of incorporating exogenous DNA, similar to other bacteria. It was thought that through this mechanism, called transformation, their inability to synthesize nucleotides could be compensated for. Mougous’s group used this competence to introduce into Southlakia a cassette with a hygromycin resistance gene, a lethal antibiotic for Southlakia but not affecting Actinomyces. The cassette also included genes encoding different fluorescent proteins. This allowed them to select hygromycin-resistant strains of Southlakia that expressed fluorescent proteins. This made it possible not only to visualize Southlakia attached to the surface of host bacteria but also to observe two very different behaviors. Many of them remained adhered without any change, but others, called “productive,” grew and produced small buds that were released, giving rise to free individuals (around 10 descendants in 20 hours). This can be seen in this video produced by Mougous’ laboratory. It was very interesting to find that Actinomyces carrying non-productive Southlakia divided and proliferated normally, but growth stopped or even reversed when carrying one or more productive Southlakia (Figure 2). This observation suggested that Southlakia growth was at the expense of molecules necessary for Actinomyces growth, indicating a parasitic relationship.
In a second set of experiments, Mougous’ group investigated which genes were essential for Southlakia life cycle. It is important to note that the phylogenetic distance between patescibacteria and other bacteria is so large that predicting the functions of their proteins through direct sequence comparison is very complex. The strategy involved massive mutagenesis using transposons, DNA segments capable of inserting into different locations within the genome. Around 40,000 insertion sites were generated, allowing for the identification of 295 genes essential for Southlakia survival. As expected, these genes are mainly related to Southlakia lifestyle and their relationships with the host, rather than to the usual metabolic pathways in bacteria that are lacking in Southlakia, as stated above. For example, one of the essential genes identified was related to the organization of pili, used both in bacterial motility and attachment to the host. Other gene was part of a secretory system essential for its survival, and another was involved in the synthesis of extracellular polysaccharides. Interestingly, the transformation systems, related to the incorporation of exogenous DNA, were found to be non-essential, suggesting that they do not represent a source of nucleotides for Southlakia DNA synthesis.
The work of Mougous and collaborators represents a crucial advance for future studies of patescibacteria, especially in understanding the genotype-phenotype relationships, as it has provided new tools for directly intervening in the genomes of these microorganisms. From now on, microbial dark matter will be a little less obscure.
The author thanks Prof. Antonio de Vicente (Dpt. Microbiology, UMA, Málaga) for his valuable comments and suggestions.
References
- Ji Y, Zhang P, Zhou S, Gao P, Wang B, Jiang J. 2022. Widespread but poorly understood bacteria: candidate phyla radiation. Microorganisms. 10, 2232. doi: 10.3390/microorganisms10112232 ↩
- Collins AJ, Murugkar PP, Dewhirst FE. 2021. Establishing stable binary cultures of symbiotic Saccharibacteria from the oral cavity. J Vis Exp. 13. Doi: 10.3791/62484. PMID: 33938896. ↩
- Wang Y, Gallagher LA, Andrade PA, Liu A, Humphreys IR, Turkarslan S, Cutler KJ, Arrieta-Ortiz ML, Li Y, Radey MC, Cong Q, Baker D, Baliga NS, Peterson SB, Mougous JD. 2023. Genetic manipulation of Patescibacteria provides mechanistic insights into microbial dark matter and the epibiotic lifestyle. Cell. 186:1-15. Doi: 10.1016/j.cell.2023.08.017. ↩