Sex and the colony
Sex and the colony

When the general media report news about genetics, there is a deplorable tendency towards gene determinism. Headlines in the vein of “Gene for X discovered”—where X is anything from a physical trait to a complex behavior—are regrettably common, and almost always incorrect. That’s why I face this article with some trepidation.
Because, you see, there is a gene that explains social structures in ants. Or rather, a supergene. But let’s begin at the beginning.
The fire ant Solenopsis invicta, apart from having the most badass name in the whole history of every ant ever, comes in two flavors of social structure. It can either have a single, large queen that will kill any other would-be queen that appears, in which case we’re talking about a monogyne social structure, or it can have several, smaller queens that coexist more or less peacefully, making it a polygyne social structure.
What determines whether a S. invicta colony will have a polygyne or monogyne social structure? Here’s a clue: workers belonging to a monogyne family will have only one allele of the Gp-9 gene. Specifically, they will all be Gp-9BB. If the allele Gp-9Bb is present in the family, then you have several queens: a polygyne social structure. Gp-9bb ants do not survive enough to reproduce 1.
So, can a single gene determine how the whole colony will be structured?
Gp-9 is a gene that codifies for an OBP: an odorant-binding protein (here I go again with smells). Odors are important to ants: they transmit information and play a role in the social organization of the colony. Workers use the different odours that Gp-9BB and Gp-9Bb queens produce to differentiate between the two.
However, monogyne and polygyne colonies differ in more than the way the chemical information is processed: male spermatogenesys and female size and fecundity are also different, and it seems unlikely that Gp-9 controls those traits too.
Turns out that Gp-9 carries along a lot of genetic baggage: it maps within a very large region of chromosome S (for Social) that forms a linkage group. Wait a sec: a linkage group?
Yes. Forget fire ants for a moment; let’s talk about sex.
Sexual reproduction is based (very roughly) on “shuffling” the genes provided by the two gametes that come together at fecundation. This shuffling is called recombination. The DNA within genes themselves does not get recombined (otherwise it would be a catastrophe), but just the genes, as units. But there are some genes that always go together. As an analogy, imagine a deck of cards imperfectly shuffled: some cards will stay together through the shuffle. Back to the genome, when this happens to two or more genes (because they’re very close together, for example), they form what’s called linkage groups. If there’s selective pressure to keep those genes together down the generations—in other words, if recombining those genes apart would be disadvantageous—, then we’re talking about a supergene. Sex chromosomes behave like that, and that’s why you have, in many organisms, some traits linked to sex (think calico cats, or daltonism in human males).
Okay, that’s enough sex talk. Back to the fire ants.
It makes sense that Gp-9, a single gene, wouldn’t be enough to account for all the characteristics of the two different social forms of the fire ants. And Wang et al., in a letter to Nature (Nature 493, 664–668) 2, reported a very elegant series of experiments in which they showed that Gp-9 is part of a supergene that explains the differences between polygene and monogyne colonies.
They used a technique called RAD sequencing 3, which can be used to find out if a region has reduced recombination, that is, if it can behave like a supergene. They wanted to know if the Gp-9 gene was in such a region. The answer was yes.
Correction: the answer was an interesting yes.
When they did the experiment in ants from a monogyne colony, which only have the Gp-9BB allele, they found 16 linkage groups. Solenopsis invicta has 16 chromosomes. A chromosome is, by nature, a linkage group. So far, so good.
When they did the same experiment in polygyne colonies ants (which also have the Gp-9Bb allele, remember), they found 15 linkage groups analogous to the chromosomes, just like the Gp-9BB monogyne colonies, and one particular linkage group in the S chromosome, precisely where the Gp-9 gene was located. This region was not a full chromosome, but it didn’t recombine either. And it is huge: it covers 12,7 Mb, more than half of the S chromosome. Moreover, it is not present in ants from monogyne colonies, where the Gp-9Bb allele is not present.
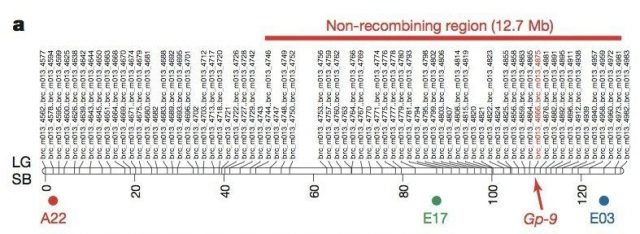
Although the whole region has not been sequenced, there are a few interesting things that point towards it being instrumental in the social differences between monogyne and polygyne colonies. First, the Gp-9 gene is there. Second, there are other 27 genes that are expressed differently between monogyne and polygyne workers, and of these, 19 can be found inside this large non-recombining region. Genes that are also expressed differently in males and queens can be found either in the supergene or in other linkage groups for preference (meaning, recombination for these genes is deactivated). Let’s see it at a glance:
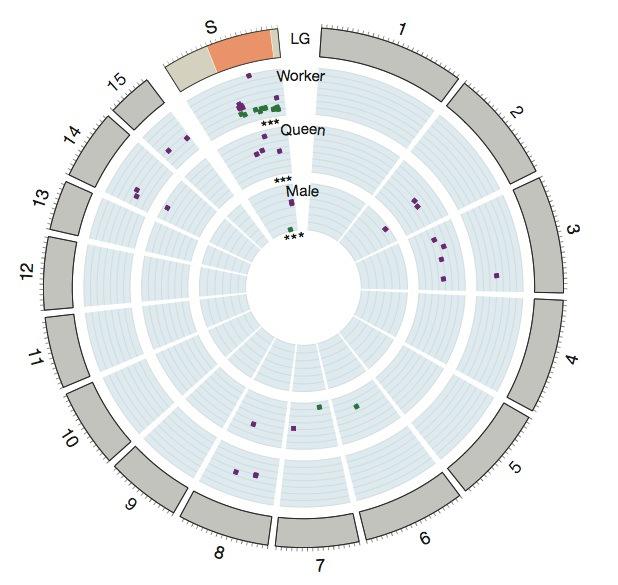
What does this mean? Supergenes are not rare. They are known to be responsible for mimicry in certain buterflies and for heterostyly (different morphological types of flowers) in primroses. These are, however, direct biological results of the phenotype. The work of Wang et al. goes a step beyond in establishing a tantalizing link between genetics and behaviour in social insects; a “social chromosome”, as they call it, that behaves in a manner remarkably similar to sex chromosomes. And since many species have both monogyne and polygyne colonies, it would be interesting to know whether an analogous of S. invicta’s social chromosome can be found in them too.
References
- Andrew F. G. Bourke & Judith E. Mank Genetics: A social rearrangement Nature 493, 612–613 (31 January 2013) DOI:10.1038/nature11854 ↩
- Wang, J. et al. Nature 493, 664–668 (2013) ↩
- Baird NA, Etter PD, Atwood TS, Currey MC, Shiver AL, et al. (2008) Rapid SNP discovery and genetic mapping using sequenced RAD markers. PLoS One 3: e3376 ↩
3 comments
VERY interesting. And you made it very easy to understand, too! Thanks! 🙂
Thanks a lot! I tried. I’m very happy you liked it!
Excellent explanation!