The Naked Brain
The human brain is arguably the most fascinating organ found in nature. Its complexity has been known since Santiago Ramón y Cajal pioneering work showed the central nervous system (CNS) was a contiguous neuronal network where neuron is the basic unit as opposed to the view of the brain being a single continuous network (reticulum) proposed by others at the time. Still, unequivocal experimental proof corroborating his findings had to be on hold until 1950s when electron microscopy was established and the existence of synapses could be finally confirmed. This anecdote just tells us how important is technological progress to get a better understanding on how brain works, but development of new methodologies to gain detailed information is key too. How could otherwise Ramón y Cajal have made his paramount observations without Camilo Golgi’s silver staining technique?
Neuroscience has made giant steps since the neuron doctrine was born and even though the brain is still puzzling to us as a whole, we do know a lot more about the physiology of its building brick, the neuron. However, the study of the entire mammalian brain (from a neuroanatomical point of view) largely relies on error-prone methods involving sectioning and reconstruction, which are tedious and often lead to misinterpretation of structural and molecular data. Effort has been made in order to study whole mouse brain including 3D imaging after tissue clearing, but these methods do not allow intact-tissue molecular phenotyping (at the protein level) and are quite lengthy (even up to weeks of tissue clearing).
Last year though, a Nature paper from Karl Deisseroth group (well known for exploring brain function by using optogenetics), gave birth to a new method called CLARITY (quite revealing) that substantially improves previous attempts to get transparent intact tissue 1. They have been able to transform mouse brain tissue into an optically transparent and macromolecule-permeable one while keeping its native molecular and structural arrangement. To do so, they create a polimerized hydrogel mesh starting from hydrogel monomers (acrylamide and bisacrylamide), folmadehyde and thermally triggered initiators. The hydrogel-tissue complex comprises the scaffold that physically supports tissue structure and preserves molecule arrangement. More important is the hydrogel mesh does not bind lipids and biomolecules lacking functional groups that can conjugate, because these molecules obstruct access to both molecular probes and photons. Removal of lipids will prevent scatter of light, which is key for quality 3D imaging and immunohistological analysis.
But how to remove lipids in an effective time frame without affecting receptors and ion channels (present in the membrane) or synapses? By using the detergent of choice in most electrophoretic techniques since decades: the ionic detergent sodium dodecyl sulphate or SDS. Application of an active-transport organ-electrophoresis in which an electrical field is applied to the already hybridized intact tissue in the presence of SDS micelles (electrophoretic tissue clearing or ETC) allowed an accelerated extraction of membranous lipids (days versus weeks when compared to passive clearing) while preserving molecular-cellular integrity.

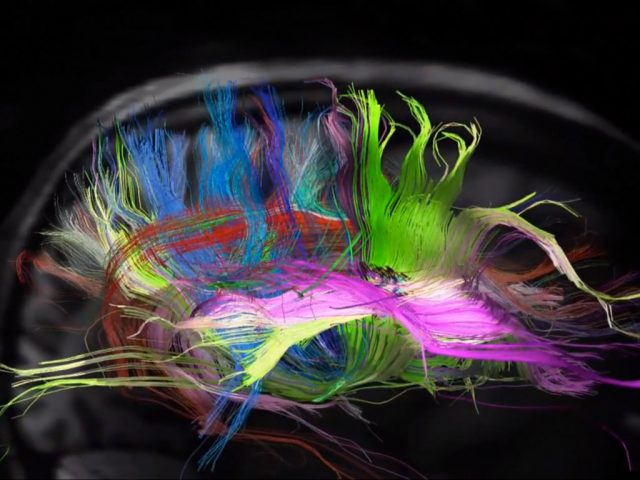
When CLARITY is applied to mice brain tissue in which the cytosolic fluorescent protein enhanced YFP is selectively expressed in a subset of neurons and the tissue is immersed in a refractive-index-specified solution matching the CLARITY hybrid composition, the intact brain becomes transparent and could be imaged at cellular resolution even using single-photon microscopy (Figure 1). Expression of YFP enabled them to identify and image these neurons within the intact brain in regions like the Thalamus or the Hippocampus with unprecedented resolution (even at 3.6 mm depth, working distance limit of the objective). In addition CLARITY involves minor protein loss while allowing several rounds of antibody labelling with little antigen damage and high penetration rate, thus making possible molecular phenotyping without sectioning. When performing multiple rounds of labelling, the ionic detergent SDS can be re-used to denature and disrupt antibody binding before application of the next antibody. In summary, technical features suggest CLARITY is the beginning of a new and improved way to look at neuroanatomy, but why CLARITY may be of value to research?
Besides the possibility of applying the technique to any tissue, as shown by a recent publication in Cell journal describing an improved method allowing mice whole-body clearing 2, the technique has been successfully used in a study aiming to disclose the neural mechanisms accounting for the robust long-lasting memories contributing to develop drug addiction (using cocaine as the substance of choice). They found recruitment of a small proportion of neurons in the lateral nucleus of the amygdala (preferentially with increased transcription factor CREB expression) after repeated cocaine use in mice (cocaine conditioned behaviour). In essence, they suggest such neural mechanisms (drug addiction) are the consequence of connections and cross talk between different regions of the brain rather than the activity in a single, predominant area 3.
Likewise, CLARITY is useful when tested in post-mortem human brains. A major problem of tissue being stored for years relies on its extensive crosslink as a consequence of formalin-mediated fixation, meaning those samples could be not suitable for molecular studies any longer. Surprisingly, application of CLARITY to the frontal lobe from an autistic patient stored for more than 6 years allowed them for immunohistological visualization and identification of neurons and projections. Tracing of individual fibres was possible after axon staining with neurofilament protein and myelin basic protein. Thereafter parvalbumin staining (present in GABAergic interneurons) was used to trace and look at the distribution of parvalbumin-reactive neurons in the neocortex over large volumes (1). Many of these interneurons, particularly in deep layers, showed abnormal isoneural and heteroneural dendritic bridges compared to age and sex matched normal brain (the way dendrites establish coverage of a territory by following the organizing principles of self-avoidance and tiling). Instead, this arborisation pattern is associated with mutations in the Down syndrome cell-adhesion molecule protein (Dscam) or protocadherins, which have been, in turn be linked with autism-spectrum disorder. This observation illustrates how informative clarified tissue may be. Its application can provide valuable molecular data of neuropsychiatric disease when comparing for instance, abnormal brain tissue from clinical brain banks (like the MRC London neurodegenerative diseases brain bank) to control tissue brain from post-mortem healthy individuals.
In conclusion, even though CLARITY is a method still on its infancy and will need further improvement, it looks one with pretty much potential. Expanding research areas such as connectomics will undoubtedly benefit from its progress making the titanic task of creating maps of connections from the nervous system possibly a little simpler.
References
- Chung K, Wallace J, Kim SY et al: Structural and molecular interrogation of intact biological systems. Nature 2013; 497: 332-337. ↩
- Yang B., Rajan P. Kulkarni, Benjamin E. Deverman, Chun-Kan Chen, Eric Lubeck, Sheel Shah, Long Cai & Viviana Gradinaru (2014). Single-Cell Phenotyping within Transparent Intact Tissue through Whole-Body Clearing, Cell, 158 (4) 945-958. DOI: http://dx.doi.org/10.1016/j.cell.2014.07.017 ↩
- Hsiang HL, Epp JR, van den Oever MC et al (2014) Manipulating a “cocaine engram” in mice. J Neurosci34: 14115-14127. ↩