Lung cancer can be detected through your nose

There is a word everybody fears and that is because when you listen to it out loud, it could be too late. Even though we have likely coexisted with cancer since complex multicellular organisms evolved, to us human beings, it just seems the number of cases keeps growing every year. Sadly, that seems to be the case at least in United Kingdom, where the number of affected individuals registered on the National Cancer Registry Database has risen by almost 200% on the period 1975-2010. Regardless of the reasons explaining such increase, the good news is physicians can cure a good proportion of patients, but only when detection happens at an early stage. Is therefore important to screen individuals who are at risk of developing certain types of cancer, but keeping in mind the financial constrains national health systems constantly face (meaning we need to look for individuals falling into the high-risk population).
A well-grounded example supporting this clinical approach came from the National Lung Cancer Screening Trial (NLST) aiming to compare two ways of detecting lung cancer and published back in 2011 1. Over a 21 month long period, 53454 heavy smokers (current or former) were split into 2 groups, one having computed tomography (CT), which consists of an imaging technique allowing to create 3D images from internal organs, and the another one having standard chest X-ray (1). The first group of individuals showed nearly 20% relative reduction in mortality associated to lung cancer, thus preventing many unnecessary (and costly) deaths over a 6.5 years of follow-up. Furthermore, a large proportion of individuals showing a pulmonary lesion ended up being diagnosed as benign nodules, meaning some patients could have avoided invasive procedures. Hence, it would be desirable and cost-effective finding predictive ways to identify patients at risk.
Professor Avrum Spira and his team at Boston Medical Campus, have made big efforts trying to characterize the so called “airway gene-expression signature” which seems to appear in a proportion of smokers. If confirmed, this genetic fingerprint could be a predictive marker because the hypothesis is lung cancer is a late event in the chain, while the “signature” is a consequence of tobacco smoke exposure and happens much earlier. Furthermore, those changes take place not just within the bronchial epithelium (more difficult to reach), but also in the more accessible nasal epithelium. This particularity would offer great advantage, as it may allow detection of high-risk individuals by collecting a sample directly from the nose, without using the more invasive and time-consuming bronchoscopy or CT techniques.
To confirm the former, they studied 554 nasal samples from two multicentre prospective studies (AEGIS-1 and AEGIS-2) that recruited patients from 28 different sites (USA, Canada and Ireland) with suspect lung cancer 2. First, they identified 535 genes differentially expressed in the nasal epithelium of lung cancer patients compared to those with benign disease (after 1 year follow-up). Second, they compared the expression pattern between nose and bronchial epithelium from a subset of patients in AEGIS-1 where microarray data was available. A significant concordance between samples could be confirmed (p-value smaller than 0.001) in both upregulated and downregulated genes from patients with lung cancer 3. Both types of airways show therefore similar differences in lung cancer-associated gene expression.
Quite remarkable was the overwhelming number of genes downregulated (492 genes) as compared to the upregulated ones (43 genes). Genes involved in processes such as DNA damage, regulation of programmed cell death or apoptosis (critical to remove potentially harmful cells from the organism) or pathways involved in immune system activation fell into the downregulated group, whereas genes involved in ion transport (including magnesium and release of calcium into the cytosol) and endocytosis were upregulated.
To evaluate whether nasal gene expression profile is a good lung cancer predictor, they selected the 30 most differentially expressed genes (from the total of 535 cancer-associated genes) achieving maximal performance in cross-validation (a predictive modelling technique used for biomarker validation) and combined this data with a clinical risk factor model. The model was based on a previous study by Gould et al. including parameters such as patient age, smoking status, time since quit and nodule size. Assessment of this combined model on 130 nasal samples from the AEGIS-2 clinical trial that were not used to develop the classifier significantly improved the sensitivity and negative predictive value for detecting lung cancer when compared to the clinical model alone (without the biomarker). This result shows for the first time that nasal gene expression can indeed be valuable to detect lung cancer and may allow physicians to perform invasive procedures only in those patients were the likelihood of having disease is high. In addition, it may allow application in circumstances where bronchoscopy is not useful because of the location of the mass or the risk of having complications.
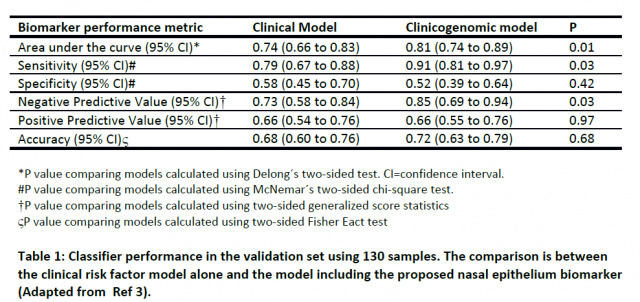
In summary, and weighting on limitations such as the use of a cohort enriched with patients having large nodules (and therefore more likely to have lung cancer) or the limited ability to assess the performance of the biomarker on the clinically relevant set of patients with small and/or peripheral nodules, the present study suggests that nasal samples may represent a new and meaningful clinical tool to effectively identify patients having malignant lung cancer growth.
References
- National Lung Screening Trial Research Team., Aberle DR, Adams AM, Berg CD,Black WC, Clapp JD, Fagerstrom RM, Gareen IF, Gatsonis C, Marcus PM, Sicks JD. (2011) Reduced lung-cancer mortality with low-dose computed tomographic screening. N Engl J Med. 2011 Aug 4;365(5):395-409. doi: 10.1056/NEJMoa1102873. ↩
- Silvestri GA, Vachani A, Whitney D, Elashoff M, Porta Smith K, Ferguson JS, Parsons E, Mitra N, Brody J, Lenburg ME, Spira A; AEGIS Study Team. (2015) A Bronchial Genomic Classifier for the Diagnostic Evaluation of Lung Cancer. N Engl J Med. 2015 Jul 16;373(3):243-51. doi: 10.1056/NEJMoa1504601. ↩
- AEGIS Study Team. (2017) Shared Gene Expression Alterations in Nasal and Bronchial Epithelium for Lung Cancer Detection. J Natl Cancer Inst. 2017 Jul 1;109(7). doi: 10.1093/jnci/djw327. ↩