Self-knotting bionic proteins
Author: Ivan Coluzza is an Ikerbasque Research Professor at CICbiomaGUNE
Some heteropolymers can be designed to spontaneously self-assemble in complex pre-determined knotted structures. In the past, we referred to such designable heteropolymers as “bionic proteins”. Here we present an extensive study on self-knotting bionic proteins.
As our everyday experience with ropes teaches us, knots form spontaneously in polymer chains of sufficient length and flexibility have interesting applications. There is no reason not to speculate that several applications for knots could be found also at the molecular or microscopic scale. In fact, it has been shown that the presence of knots also at these scales, such as in proteins or artificial polymers, can enhance mechanical and thermal stability. This stabilisation of the polymer structure could have interesting applications in engineering cargo-carrier constructs for instance for the delivery of drugs in the body, as the stability of the structure of the cargo is essential to ensure the success of the delivery. Moreover, knots seem to play a role in proteins that act as catalysers (enzymes), i.e. proteins that accelerate natural chemical reactions in the body. Therefore knots could have a significant effect on catalytic activity also in synthetic polymers.
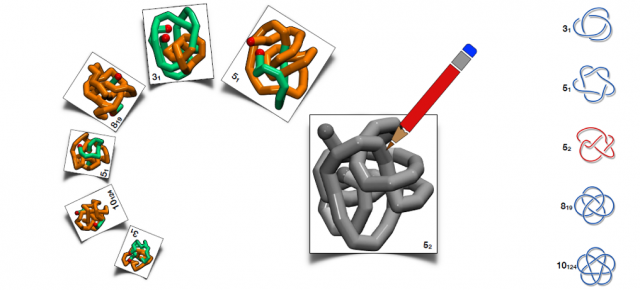
However, to this date, the synthesis of a chosen knot is still a challenging task. Most of the progress in this direction is represented by little knots, engineered via a chemical synthesis method that is able to synthesise the simplest knot (so-called trefoil knot), and only recently a few other more complex knot types. Of the billions of types of knots existing, only four have been selectively manufactured using small building blocks, making the synthesis of specific knots still an open problem. In this manuscript 1, we present a significant advancement to the solution of this problem, showing that polymers composed of chains of different monomers (heteropolymers) can be designed to self-assemble into specific chosen knot types with a precise structure, by choosing the right sequence of different monomers along the chain. These chains can be then locked externally by closing the ends of the polymer in order to lock the knot, and this has been shown to bring thermal and mechanical stability of the structure.
In the design of knotted synthetic heteropolymers, the state of the art is still limited to design of sequences that enhance the total knotting probability, without being able to create a specific knot. Heteropolymers are molecular chains that model polymers made by a different alphabet of building blocks. Each monomer type interacts with the other categories according to a generalised interaction matrix qXq where q is the alphabet size. The results presented here do not depend on the specific value of the interaction provided that the average value and the spread of the values in the matrix are conserved. A heteropolymer consists then into a sequence of q types of monomers, and the sequence encodes for the function of the chain as we demonstrated in several previous works. Proteins inspired our work as they are the best-known example of natural heteropolymer.
We have considered the alphabets q = 20 (same as proteins) and q = 3 (a small alphabet we proved to be sufficient for the encoding). We employed computer modelling to explore the vast space of chain conformations and sequences. The computer tries to modify either the shape of the chain or mutate the sequence, the change it is then accepted or rejected if it is compatible with the thermal fluctuations at equilibrium.
Through our computational study, we observe a broad variety of knots of complexity up to more than 14 crossings, we design heteropolymers that self-assemble with high precision in all knot types experimentally constructed up to now, plus also an unprecedented twist knot (type 52) and the exotic “pretzel knot” with 10 crossings (type 10124). Thus, our design strategy can be used as a guideline for the experimental construction of more complicated knots.
Our study also has important implications in our understanding of protein evolution. An open question in structural biology is related to the relatively low abundance of knotted proteins compared to what is found in conventional polymers and other biopolymers (e.g. DNA/RNA). Previous studies have speculated the low number of knotted natural protein structures is the result of an active selection of evolution over the sequence of the protein to avoid knots. In this manuscript instead, we show that the increase in the number of different monomers (alphabet size) spontaneously drives the system to reduce the complexity of the knots, or eliminate them altogether. Hence, with respect to DNA or RNA that have an alphabet size of 4, the large alphabet of proteins (composed by 20 monomers, the amino acids) has the property of lowering the spontaneous occurrence of knots without losing the possibility of optimising complex knots, a hypothesis that was never speculated before.
References
- Chiara Cardelli, Luca Tubiana, Valentino Bianco, Francesca Nerattini, Christoph Dellago, and Ivan Coluzza (2018) Heteropolymer Design and Folding of Arbitrary Topologies Reveals an Unexpected Role of Alphabet Size on the Knot Population Macromolecules doi: 10.1021/acs.macromol.8b01359 ↩