T-cell acute leukaemia exhibits dynamic interactions with bone marrow microenvironment
T-cell
Author: Marta Irigoyen is a postdoctoral researcher at CIC bioGUNE
T-ALL is an aggressive malignancy which results from the leukemic transformation of T-cell progenitors into tumor cells. It is widely accepted that complex interactions between T-ALL cells and their surrounding microenvironment contribute to disease and may regulate quiescence, survival and self-renewal of cancer cells themselves. This relationship may underlie a critical mechanism that facilitates both the initiation of disease and chemoresistance.
Leukaemia develops within the bone marrow (BM), where it has been suggested to take part in a complex crosstalk that in some cases results in microenvironment remodelling 12. Since therapeutic targeting of leukaemia-supportive niches has been proposed 34, is critical to understand both the spatial and kinetic nature of leukaemia-BM interactions. However, the current knowledge of leukaemia biology is predominantly derived from ex vivo analysis and static images that cannot capture information on the location and dynamics of leukaemia interactions with BM structures and cells over time.
To address these questions, Hawkins ED and coworkers 5 monitored leukaemia growth in mouse bone marrow by intravital microscopy (an optical imaging technique to observe living organisms in vivo at microscopic resolution through windows done on the tissue of interest) 67. To characterize T-ALL interactions in vivo, they injected leukaemic cells and showed that T-ALL preferentially infiltrated the BM before expansion to peripheral lymphoid organs. They visualized that T-ALL in the BM is randomly distributed relative to osteoblasts (bone cells responsible for synthesizing bone matrix), vasculature and perivascular mesenchymal stem/progenitor cells during disease progression.
To identify the mesenchymal stem/progenitor cells they identified the presence of the nestin intermediate filament characteristic of stem and progenitor cells cytoskeleton tagging as “nestin+” cells. Furthermore, to recognize and label these cells they used molecular genetical engineering modifying these cells to express a jellyfish fluorescent protein (GFP) tagged to the nestin (becoming “nestin-GFP+” cells becoming visible as green color cells under fluorescence microscope).
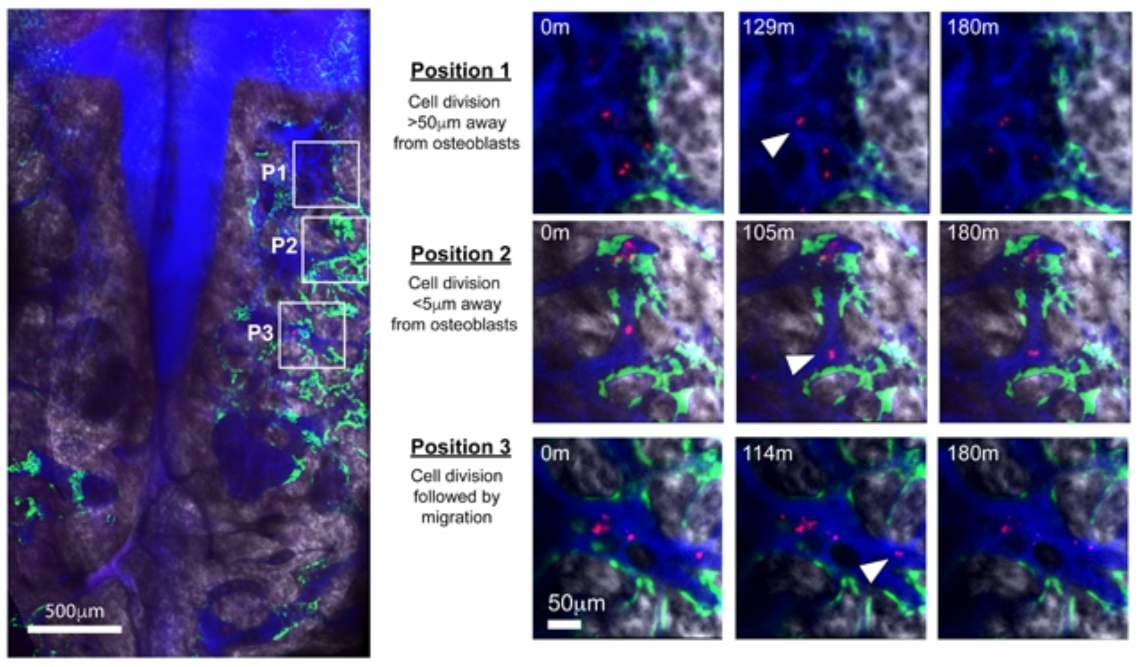
To determine whether T-ALL expansion was supported by specific constituents of the BM, they monitored the dynamics of single T-ALL cells and this revealed that the vast majority of T-ALL cells were motile and that movement was rarely restricted to the proximity of any specific cell types or structures. Notably, the speed of any given cell over time was also heterogeneous, and thus no single migratory behavior was associated with osteoblastic, nestin-GFP+ cells or vasculature. Tracking single T-ALL cells revealed the same random distribution of dividing cells and suggesting that proximity to these stroma components is not key for T-ALL expansion (stroma is extracellular matrix that the cells create to help them with nutritional support, removal of waste products and inter-cellular signaling). Additionally, daughter cells migrated large distances following division, showing that clones and their progeny are not restricted to foci within the local microenvironment (foci is the plural of focus and implies only microscopic visualization of the tumor cells), demonstrating that T-ALL seeding and colonization of BM do not depend on specific BM stroma.
The question remained whether certain BM regions could create “hotspots” for chemo-resistance through provision of a protective environment. To address this issue, they adapted their imaging protocol to follow the same BM areas over multiple days to track leukaemia dynamics from complete BM infiltration and throughout therapy. Dexamethasone, a chemotherapeutic agent in the treatment of ALL, was administered daily and immediately after the third therapy dose they observed an amazing reduction in disease burden. Surprisingly, surviving cells were highly migratory and were founded dispersed throughout the BM space and did not maintain long-lasting associations with osteoblastic or nestin-GFP+ cells.
Collectively, these data oppose the prevailing hypothesis that therapy-resistant leukaemic cells depend on a particular microenvironment for survival. To investigate this proposed paradigm, they compared the gene expression profile of T-ALL cells purified at full infiltration to those isolated from mice 7-10 days after initiation of dexamethasone treatment, when surviving cells have recolonized the BM. The gene analysis of resistant cells overlapped with that of T-ALL cells pretreatment. Indeed, only 79 genes were differentially expressed in the post-treatment group and none of the differentially expressed genes were related to known cell-niche interaction candidates. Together, their imaging and gene expression data suggest that dexamethasone treatment does not select for a subpopulation of T-ALL cells that have been directed to a specific niche.
Finally, to assay whether T-ALL may affect BM structures, they performed time-lapse imaging in heavily disease-burdened, untreated mice. They observed remarkable remodeling of osteoblasts which underwent apoptosis. However, nestin-GFP+ cells were maintained, and blood vessels could still be visualized.
In summary, the authors demonstrate that T-ALL infiltrates the BM and survives chemotherapy independently of stable interactions with specific microenvironment. Thus, their results suggest that to avoid the development of chemo-resistance, novel therapeutic interventions may not need to target specific BM stroma components, but rather the ability of T-ALL cells to interact with and migrate through the BM.
References
- Duan CW, Shi J, Chen J, Wang B, Yu YH, Qin X et al. Leukemia propagating cells rebuild an evolving niche in response to therapy. Cancer cell 2014; 25: 778-793. PMID: 24937459 DOI: 10.1016/j.ccr.2014.04.015. ↩
- Schepers K, Pietras EM, Reynaud D, Flach J, Binnewies M, Garg T et al. Myeloproliferative Neoplasia Remodels the Endosteal Bone Marrow Niche into a Self-Reinforcing Leukemic Niche. Cell Stem Cell 2013; 13: 285-99. PMID: 23850243 DOI: 10.1016/j.stem.2013.06.009. ↩
- Jin L, Tabe Y, Konoplev S, Xu Y, Leysath CE, Lu H et al. CXCR4 up-regulation by imatinib induces chronic myelogenous leukemia (CML) cell migration to bone marrow stroma and promotes survival of quiescent CML cells. Mol Cancer Ther 2008; 7: 48-58. PMID: 18202009. DOI: 10.1158/1535-7163. MCT-07-0042. ↩
- Weisberg E, Azab AK, Manley PW, Kung AL, Christie AL, Bronson R et al. Inhibition of CXCR4 in CML cells disrupts their interaction with the bone marrow microenvironment and sensitizes them to nilotinib. Leukemia 2012; 26: 985-990. PMID: 22182920. DOI: 10.1038/leu.2011.360. ↩
- Hawkins ED, Duarte D, Akinduro O, Khorshed RA, Passaro D, Nowicka M et al. T cell acute leukaemia exhibits dynamic interactions with bone marrow microenvironments. Nature 2016; 538: 518-522. PMID: 27750279 DOI:10.1038/nature19801 ↩
- Celso CL, Fleming HE, Wu JW, Zhao CX, Miake-Lye S, Fujisaki J et al. Live-animal tracking of individual haematopoietic stem/progenitor cells in their niche. Nature 2009;457: 92-96. PMID: 19052546. DOI: 10.1038/nature07434. ↩
- Scott MK, Akinduro O, Celso CL. In vivo 4-dimensional tracking of hematopoietic stem and progenitor cells in adult mouse calvarial bone marrow. J Vis Exp 2014; 91:51683. PMID: 25225854. DOI: 10.3791/51683. ↩