The largest animal genome
The largest animal genome
Author: Ramón Muñoz-Chápuli has been Professor of Animal Biology in the University of Málaga until his retirement. He has investigated for forty years in the fields of developmental biology and animal evolution.
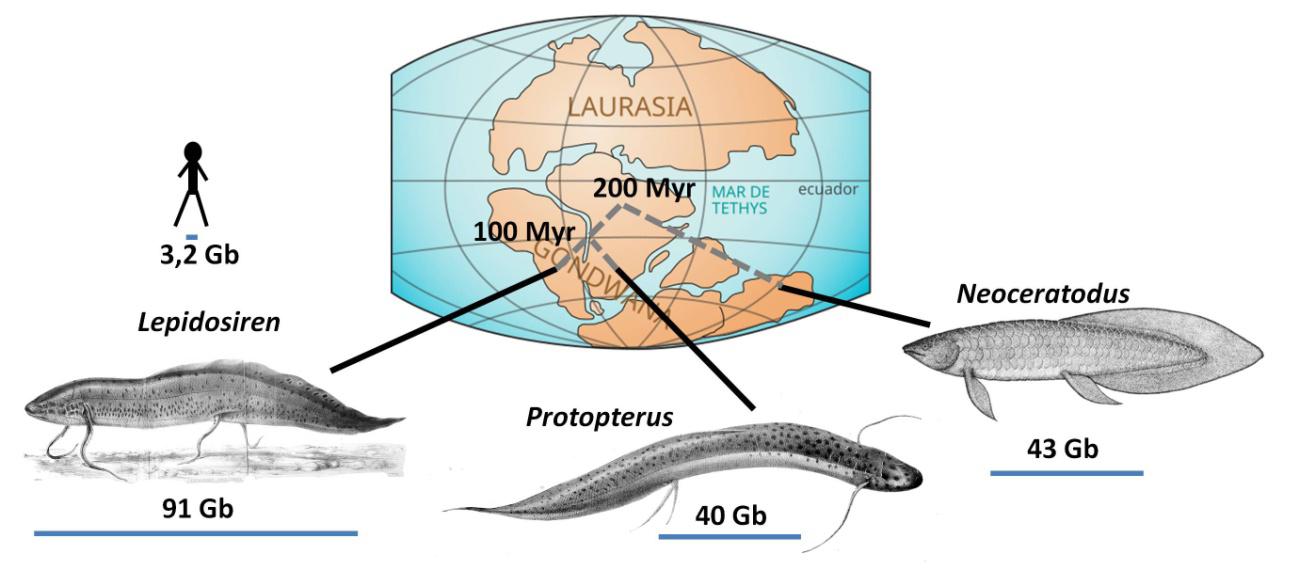
Lungfish (Dipnoi) are fascinating animals. Despite having gills, they can breathe atmospheric air thanks to a pair of lungs. Their geographic distribution aligns with the fragmentation of the supercontinent Gondwana. The Neoceratodus lineage separated from the rest 200 million years ago, becoming restricted to Australia. The ancestors of Protopterus and Lepidosiren became isolated 100 million years ago when Africa and South America split (Figure 1). The South American Lepidosiren (Figure 2) has adapted to environments where no other fish can survive. It can stay for months buried in mud, with a reduced metabolism and breathing through small orifices. And now we also know that Lepidosiren possesses the largest genome in the entire Animal Kingdom.

A study by an international team 1 has shown that the genomic DNA of Lepidosiren contains 91 billion base pairs (91 Gb). To put this in perspective, this is 30 times the size of the human genome (about 3.2 Gb). This shatters the previous records held by the other two lungfish 2, Protopterus (40 Gb) and Neoceratodus (43 Gb) (Figure 1). Among vertebrates, and with the exception of some amphibians like the Mexican axolotl (32 Gb), none come close.
A larger genome does not mean a greater number of genes. The Nature study estimates that Lepidosiren has nearly 20,000 protein-coding genes, a number very similar to that of humans. Therefore, the question is: what mechanisms have led to the accumulation of such an enormous amount of non-coding DNA in the lungfish genome? To answer this, we need to explain something about this type of DNA.
It is well known that most of our genome does not code for protein sequences. In fact, more than half is made up of sequences that are repeated more or less frequently. Although these sequences were initially dismissed as “junk DNA” we now know that they are important for genome organization and remodelling, as well as for generating mutations and increasing genetic diversity.
A key part of these repetitive sequences has the ability to replicate and move within the genome, hence they are called mobile or transposable elements. These include transposons, DNA sequences that directly “jump” from one place to another in the genome, and retrotransposons, DNA sequences that behave very similarly to retroviruses. Retrotransposons transcribe into an RNA strand that, with the help of the enzyme reverse transcriptase, reproduces the original DNA sequence, which then gets inserted elsewhere in the genome (Figure 3).
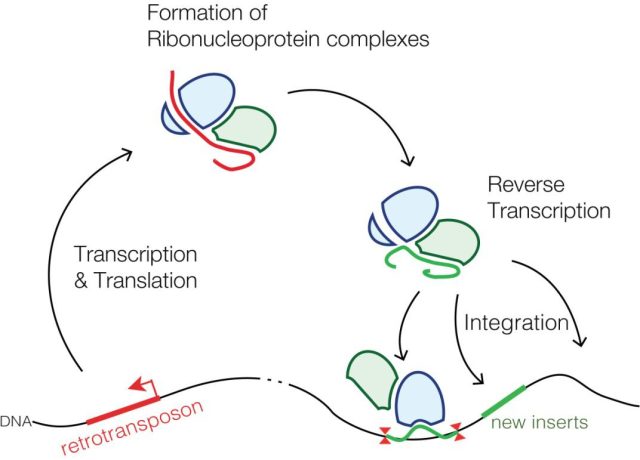
Retrotransposons are responsible for about a third of our DNA. One might think that these sequences inserting themselves into the genome could cause problems, and indeed they sometimes do. But as mentioned earlier, they also have great evolutionary significance due to their ability to generate genetic diversity, the raw material of evolution. Furthermore, there are control mechanisms that restrict the activity of retrotransposons and prevent their uncontrolled expansion. One of the most important of these is piRNA (Piwi-interacting RNA), short RNA sequences (26-32 nucleotides) that interfere with and silence retrotransposons. In fact, although retrotransposons are abundant in our genome, only about a hundred are active.
The large size of the lungfish genome is due precisely to the enormous abundance of repetitive elements in the genome, especially retrotransposons and transposons. According to the study published in Nature, this abundance correlates, among other factors, with lower control by piRNAs. These are less abundant than in other fish, and their lengths are shorter on average, reducing their effectiveness in controlling retrotransposons.
Defects in the control of mobile genomic elements have caused the Lepidosiren genome to expand at a surprising rate over the last 100 million years. It has been estimated that every ten million years, the increase in size is equivalent to the entire human genome. And the process continues today, as retrotransposon expression remains active in the Lepidosiren genome, which will continue to grow in the future.
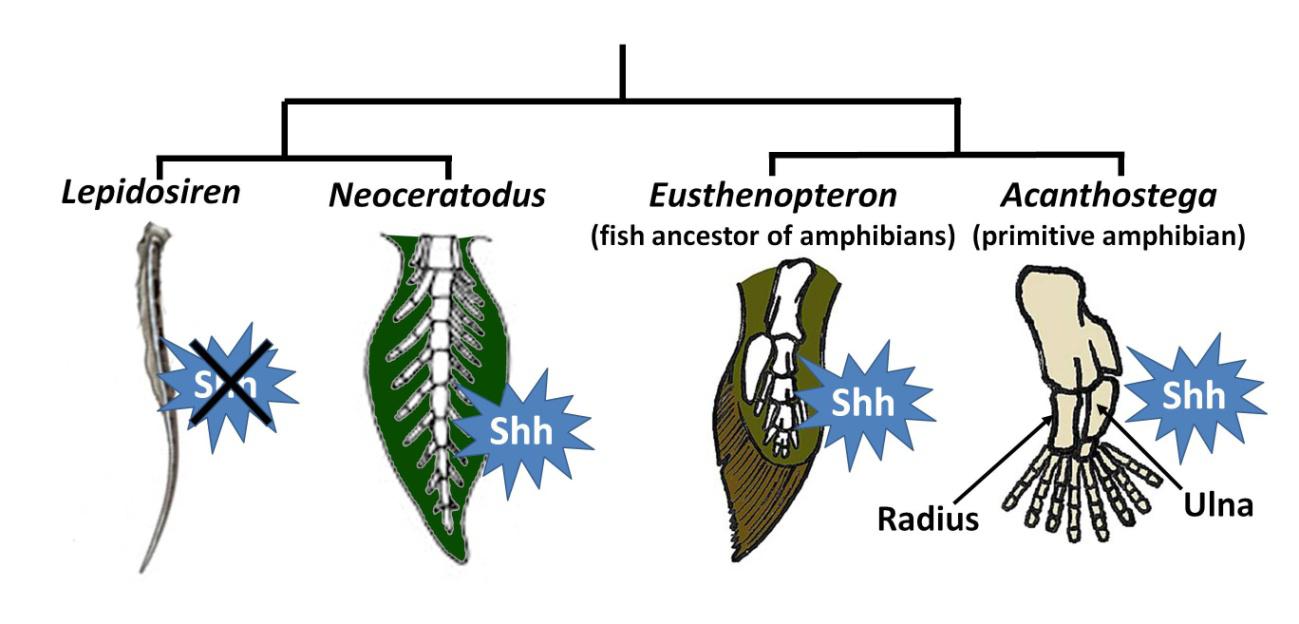
The knowledge of the Lepidosiren genome has also helped resolve an evolutionary question about the paired fins of these fish. As shown in the figure 4, the fins of Neoceratodus have a series of bony elements (radials) that resemble the ancestral fins of tetrapod limbs. In contrast, Protopterus and Lepidosiren have filamentous paired fins. It has now been shown that this difference is due to the loss of expression of a gene, Sonic hedgehog (Shh), during fin development. This gene is crucial for organizing the pattern of our fingers and toes. Its lack of expression is responsible for the absence of radials in the paired fins of Protopterus and Lepidosiren (Figure 4). In fact, in a spectacular experiment, if the fin of Lepidosiren is cut and during its regeneration is treated with a substance that acts like the Shh protein, radials are formed, reproducing the ancestral fin lost 100 Myr ago.
Apart from other surprises that the study of the Lepidosiren genome will undoubtedly bring, it will be important to understand how these animals are capable of managing such a gigantic and continually expanding genome.
References
- Schartl, M., Woltering, J.M., Irisarri, I. et al. (2024) The genomes of all lungfish inform on genome expansion and tetrapod evolution. Nature doi: 10.1038/s41586-024-07830-1 ↩
- Meyer, A., Schloissnig, S., Franchini, P. et al. (2021) Giant lungfish genome elucidates the conquest of land by vertebrates. Nature doi: 10.1038/s41586-021-03198-8 ↩