Could the largest known proteins be synthesized by the smallest bacteria?
Author: Ramón Muñoz-Chápuli has been Professor of Animal Biology in the University of Málaga until his retirement. He has investigated for forty years in the fields of developmental biology and animal evolution.
On October 11th, I published an article on Mapping Ignorance about the microbial dark matter, the vast diversity of microorganisms that cannot be cultured in the laboratory but are known through genomic and metagenomic studies of environmental samples. Specifically, we discussed the Patescibacteria, a group of extremely small bacteria with a highly reduced genome, relying on a parasitic lifestyle for survival. Today, we turn our attention to another group of peculiar components of microbial dark matter, the Omnitrophota, discovered in 1998 from genomic sequences extracted from Yellowstone Park lakes. These microorganisms are extremely small (0.2-0.3 micrometers in length) with a highly reduced genome (around 2 Mb on average, less than half the size of E. coli‘s genome).
The first complete genome information for an Omnitrophota species, tentatively named Candidatus Omnitrophus magneticus 1, was obtained in 2015. Single-cell sequencing techniques were employed for this purpose. Candidatus Omnitrophus magneticus is a flagellated bacterium, free-living, and capable of producing magnetosomes, organelles containing magnetite crystals that enable the organism to orient itself relative to the Earth’s magnetic field. However, a subsequent study on numerous Omnitrophota genomes revealed that most of them have very different characteristics, as they require living in association with other microorganisms and have parasitic or even predatory lifestyles 2.
A breakthrough occurred in 2022 when enriched media for Omnitrophota were obtained 3. It took years of cultivating Methanoseta (a genus of methanogenic archaea) to amplify the content of Omnitrophota in the culture. A presumably clonal population of Omnitrophota, named OP3 Lim, was finally isolated through centrifugation. As suspected, electron microscopy images from the cultures showed OP3 Lim cells either free or adhered to Methanoseta. Probes detecting the presence of ribosomal RNA showed three times more signal when OP3 Lim was adhered compared to when it was free, indicating an increase in its metabolism. Adhesion also increased the expression of a set of proteins, including one very large protein composed of 40,000 amino acids. Since all indications suggested that OP3 Lim cells fed on Methanoseta, leading to its death, its provisional specific name was Candidatus Velamenicoccus archaeovorus.
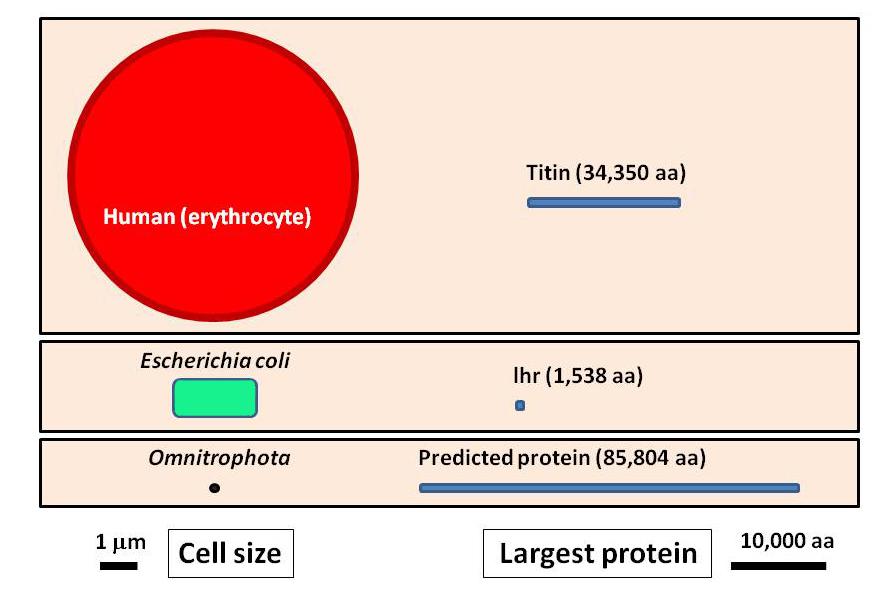
I mentioned the presence of a giant protein in these Omnitrophota, consisting of 40,000 amino acids. This is truly exceptional. To date, the largest known protein is titin, a protein in our striated muscles, with around 34,000 amino acids. The possibility of megaproteins in Omnitrophota prompted a new study, the results of which have just been released in a preprint 4. After analyzing 1800 Omnitrophota genomes, it was discovered that 46 genes encoded proteins of more than 30,000 amino acids. This implies a length exceeding 90,000 base pairs for these genes. The absolute record was a monstrous protein of 85,804 amino acids, with its gene extending along 257,000 base pairs of the bacterial chromosome. This would be the largest protein ever found, more than doubling the size of muscular titin. However, it’s crucial to note that these are predicted proteins based on gene sequences, not observed ones. In other words, it is not yet known whether the gene transcribes into a single giant messenger RNA chain, nor whether the translation of the messenger generates a single huge polypeptide chain. It could happen that either the messenger RNA or the protein is cleaved into fragments, eventually resulting in smaller proteins with different functions.
While awaiting confirmation, the study’s authors turned to AlphaFold, an artificial intelligence system developed by DeepMind (of Alphabet), to make predictions about the three-dimensional structure of the hypothetical giant protein. The system predicted domains with different functions, including some involved in cell wall adhesion, polysaccharide degradation, and a tubular structure that could be used to inject or absorb substances from prey. Thus, this hypothetical protein could play an essential role in the predatory lifestyle of these bacteria.
Confirmation of whether we are dealing with the largest known protein in one of the smallest organisms is eagerly anticipated. In any case, understanding the lifestyle of Omnitrophota will be crucial in studying the dynamics of bacterial populations and the role of ultramicrobacterial epibionts in recycling microbial biomass.
Acknowledgements
The author thanks Prof. Antonio de Vicente (Dpt. Microbiology, UMA, Málaga) for his valuable comments.
References
- Kolinko S, Richter M, Glöckner FO, Brachmann A, Schüler D. (2015) Single-cell genomics of uncultivated deep-branching magnetotactic bacteria reveals a conserved set of magnetosome genes. Environ Microbiol doi: 10.1111/1462-2920.12907. ↩
- Seymour CO, Palmer M, Becraft ED, Stepanauskas R, Friel AD, Schulz F, Woyke T, Eloe-Fadrosh E, Lai D, Jiao JY, Hua ZS, Liu L, Lian ZH, Li WJ, Chuvochina M, Finley BK, Koch BJ, Schwartz E, Dijkstra P, Moser DP, Hungate BA, Hedlund BP (2023) Hyperactive nanobacteria with host-dependent traits pervade Omnitrophota. Nat Microbiol doi: 10.1038/s41564-022-01319-1. ↩
- Kizina J, Jordan SFA, Martens GA, Lonsing A, Probian C, Kolovou A, Santarella-Mellwig R, Rhiel E, Littmann S, Markert S, Stüber K, Richter M, Schweder T, Harder J. (2022) Methanosaeta and “Candidatus Velamenicoccus archaeovorus”. Appl Environ Microbiol doi: 10.1128/aem.02407-21. ↩
- West-Roberts J, Valentin-Alvarado L, Mullen S, Sachdeva R, Smith J, Hug LA, Gregoire DS, Liu W, Lin TY, Husain G, Amano Y, Ly L, Banfield JF (2023) Giant genes are rare but implicated in cell wall degradation by predatory bacteria. Preprint in BioRxiv doi: 10.1101/2023.11.21.568195). ↩