BiaPy, analising bioimages made easy
BiaPy, analising bioimages made easy
Imagine peering through a microscope, marvelling at the intricate dance of cells, proteins, or tiny structures within a living organism. These images, known as bioimages, are a treasure trove of information for scientists studying life’s mysteries, from how cells divide to how diseases disrupt tissues. But analysing these images is no small feat. The sheer volume of data and the complexity of patterns often require advanced computational tools. Enter BiaPy 1 , a groundbreaking open-source platform that’s making bioimage analysis accessible to researchers, regardless of their coding expertise. Published in *Nature Methods*, BiaPy is a game-changer in the world of biology, blending deep learning with user-friendly design to unlock new discoveries.
Focus on science rather than coding
Bioimages, like those from electron or fluorescence microscopes, capture details invisible to the naked eye—think of mitochondria powering cells or the delicate architecture of a fruit fly embryo. Traditionally, analysing these images meant manually sifting through thousands of pictures or relying on specialized software that demanded advanced programming skills. This created a barrier for many biologists, who are experts in life sciences but not necessarily in computer science. BiaPy dismantles this barrier. It’s like giving researchers a powerful, easy-to-use microscope for data, allowing them to focus on science rather than coding.
Recognizing patterns
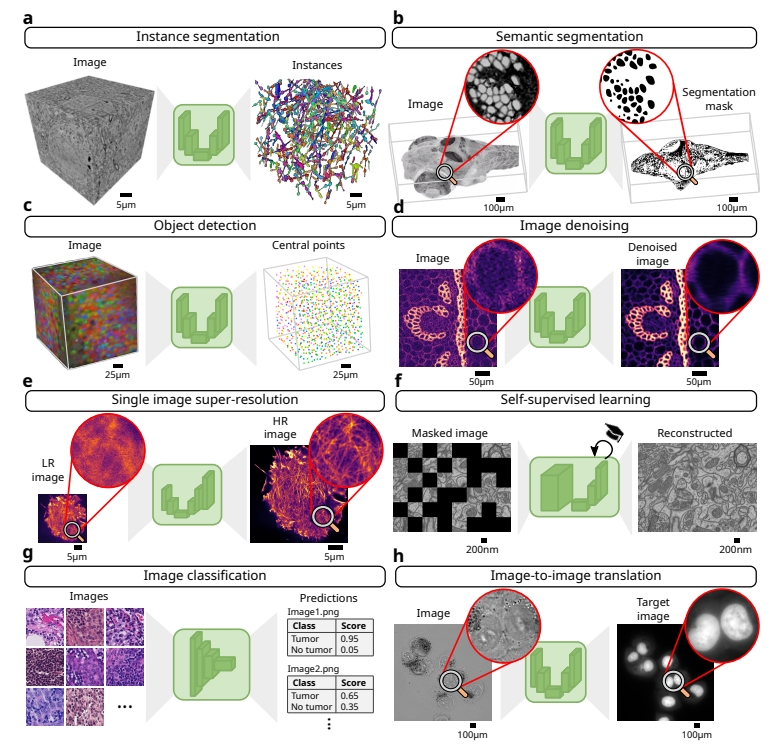
At its core, BiaPy uses deep learning, a type of artificial intelligence inspired by how the human brain processes information. Deep learning models can “learn” to recognize patterns in images, such as identifying cell boundaries or spotting specific structures, by training on example data. BiaPy supports a variety of tasks, from segmenting individual cells in 3D images to denoising blurry microscope captures or even enhancing low-resolution images to reveal finer details. What sets BiaPy apart is its versatility—it handles both 2D and 3D images, supports multiple microscope types, and works with cutting-edge models like Transformers, which are revolutionizing AI by processing complex data more effectively.
A triumph of accessibility
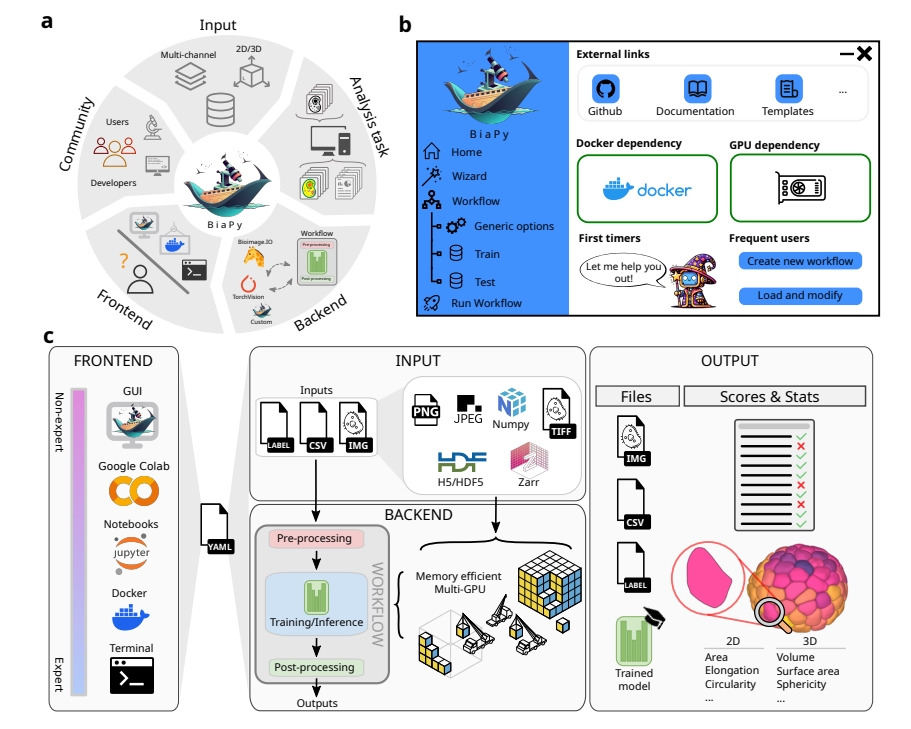
The platform’s design is a triumph of accessibility. Instead of requiring users to write complex code, BiaPy offers a graphical interface and “zero-code” notebooks, where researchers can set up analyses with a few clicks. It’s like using a smartphone app instead of programming a computer from scratch. For those working with massive datasets, BiaPy scales up, leveraging multiple graphics processing units (GPUs) to crunch data quickly. It also supports large file formats, ensuring it can handle the hefty images produced by modern microscopes. A single configuration file, editable through a text editor or guided “Wizard” tool, lets users tailor workflows to their needs, making the process intuitive and shareable.
Already in use
BiaPy’s impact is already evident in real-world research. For instance, it’s been used to segment mitochondria in large electron microscopy volumes, revealing their complex shapes in unprecedented detail. In another study, it helped model wound healing in fruit fly embryos by analyzing time-lapse videos, offering insights into how tissues repair themselves. It’s even streamlined the analysis of epithelial cysts, automating tedious tasks to uncover patterns in cell morphology. These examples show BiaPy’s power to accelerate discoveries across diverse biological questions.
Open-source tool
As an open-source tool, BiaPy invites collaboration, encouraging scientists to contribute ideas and improve its features. While it doesn’t cover every task—like image visualization, which requires other tools—it complements existing platforms, filling gaps in scalability and ease of use. Available on GitHub with detailed tutorials, BiaPy empowers researchers worldwide to harness AI without needing a computer science degree. By democratizing bioimage analysis, it’s paving the way for faster, more inclusive scientific breakthroughs, bringing us closer to understanding the building blocks of life.
Author: César Tomé López is a science writer and the editor of Mapping Ignorance
Disclaimer: Parts of this article may have been copied verbatim or almost verbatim from the referenced research paper/s.
References
- Daniel Franco-Barranco, Jesús A. Andrés-San Román, Ivan Hidalgo-Cenalmor, Lenka Backová, Aitor González-Marfil, Clément Caporal, Anatole Chessel, Pedro Gómez-Gálvez, Luis M. Escudero, Donglai Wei, Arrate Muñoz-Barrutia, and Ignacio Arganda-Carreras (2025) BiaPy: accessible deep learning on bioimages Nature Methods doi: 10.1038/s41592-025-02699-y ↩