Parasites, chromosomes and the tree of life

Advances in the knowledge of the molecular basis of life have provided some of the most impressive and certain proofs to the notion that all organisms on Earth form part of a unique and ancient lineage. One of the most typical examples is the fact that the molecular machinery regulating the biochemical processes common to the vast majority of life forms is incredibly similar, from unicellular organisms to us, pluricellular and self-conscious humans. And though finding and comparing similarities is a nice way to reconstruct genealogies, the same kind of information can be inferred when paying attention to the differences. Putting together all these similarities and differences between organisms, from the highest macroscopic features to the tiniest molecular details, we have been able to compose a rough draft of what is usually called the tree of life: the relationship between all beings that have ever lived in our planet. Since the first drawings on Charles Darwin’s voyage notebooks, to the modern phylogenetic trees produced through the power of hundreds of processing cores, this tree has grown and adopted several forms, sometimes being more similar to a huge set of bushes that arise from a common stem 1. In any case, although the main branches that connect the most obvious groups of organisms are confidentially clear, there are many gaps and cryptic relationships yet to be solved. Today, we will learn about one of the differences that may help define one of the major basal branches of this huge tree: not less than the one that defines the first eukaryotic organisms. But before we get to this epic resolution, we should first make some concepts clear.
Starring in today’s story, we have a group of unicellular flagellates called kinetoplastids. The weird name of these organisms comes from a particular structure, the kinetoplast, which is basically a granule of DNA (plus some regulatory proteins) kept within the bug’s unique and lonely mitochondrion. Kinetoplastids belong to a broader group called Euglenozoa, and many of them spend their lives being parasites of animals. In humans, several species of kinetoplastids are responsible for diseases as important as sleeping sickness or Chagas disease, for instance.
But we started talking about trees of life here; from an evolutionary point of view, there is quite uncertainty about the position of Euglenozoa within the eukaryotic lineage. All data point to a quite basal position in the eukaryotic tree, meaning that this group diverged long before other big and famous groups of organisms like plants or animals even appeared. So in the end, if all eukaryotes are relatives, then kinetoplastids are our far cousins. How far? Are they closer to us than other, really ancient, unicellular parasites? Were they the first eukaryotic organisms to split from the main stem that give rise to the other groups? Those are the key questions yet to be answered. Right now, the only concept we need to assume is that at the cellular and molecular level, we should have in common, at least, the main molecular machinery with this parasites. And here comes the surprise.
Bungo Akiyoshi and Keith Gull, from the University of Oxford 2, have just found out that all known kinetoplastids lack one of the most conserved set of proteins among eukaryotes: the ones that form the molecular macrostructure known as kinetochores, a protein scaffold that permits the correct interaction between chromosomes and the cytoskeleton during the complex process of cell division. In easy words: when an eukaryotic cell gets ready to divide into two cells, genetic information is replicated and condensed in the form of pairs of identical chromosomes, in order to make an even delivery of genetic information towards the two nascent cells. It is a process largely controlled by physical forces, in which huge molecular mobile pillars known as microtubules (these are the cytoskeleton components) push or pull each chromosome towards the pole of a gigantic double cell that soon will split into two smaller ones, each one of them carrying, in the end, a complete set of chromosomes. The interaction between the microtubule and the DNA takes place by means of a protein scaffold located in the middle section (the centromeric region) of each chromosome: that scaffold is what we call the kinetochore.
Proteins involved in the division process (cell cycle proteins, microtubule proteins, and so on) are counted among the most similar in eukaryotes. They have changed very little during the million years of separate evolution of species, because they were very fit for the purpose they had to do, and this purpose did not change from one group of organisms to another. Then, why do kinetoplastids lack the proteins that constitute a typical kinetochore? Do they also lack chromosomes? The striking fact is that they do not. Even though they exhibit particularities, they largely divide like the rest of eukaryotic cells, so if we compare the proteins usually involved in the control of cell cycle, we will find that there are kinetoplastid versions for almost all of them. The weirdness comes when analyzing the kinetochores…
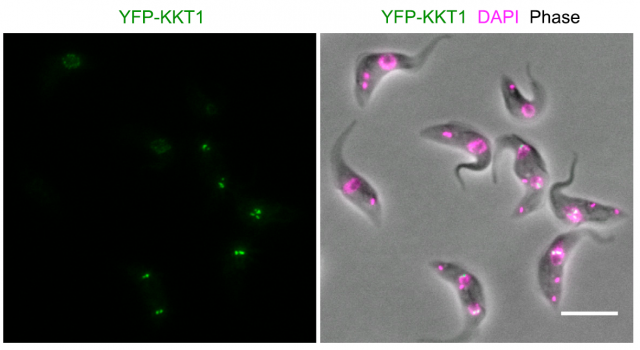
Modern bioinformatics permit an easy comparison between proteins from different organisms. Using these techniques, no similarities were found with any other known kinetochore-like protein after searching the complete genome of several kinetoplastid species. Particularly striking was the absence of a recognizable version of the ubiquous CENP-A protein, which interacts with the chromatin placed at the centromeric region of chromosomes and permits the assembly of kinetochores in almost all known eukaryots. Since finding kinetochore proteins like CENP-A by similarities with other genomes was unsuccessful, the researchers made a clever move: they sequentially tagged all proteins in the kinetoplastid genome with a fluorescent dye, and using advanced microscopy techniques, they examined the cells searching for specific patterns of fluorescence that correlated with the movement of chromosomes during cell division. In this way, they could identify 19 proteins that accomplished the criteria for putative kinetochore proteins (which they named KKT proteins, standing for kinetoplastid kinetochore). They could now sequence the amino acid composition of the 19 candidates and prepare a series of tests that further confirmed if they fit with the expected functions for a kinetochore protein. They found that most of them interacted specifically which one another, and a few of them interacted specifically with particular regions on the DNA at the centromeric region. A series of experiments based on knocking down the expression of the proteins in the kinetoplastid showed tha these parasites were not able to divide normally and their chromosomes were not correctly separated from one another into the nascent cells.
And yet, further bionformatics analysis of the amino acid composition of these novel proteins confirmed that their sequence was particularly different from the canonical kinetochore proteins. This seems to imply that the process of chromosome segregation is largely ruled by physical requirements: since the biochemical properties of kinetochore proteins can drastically vary, the most important requirement seems to be the capacity to bind the centromeric DNA and the microtubules at the same time. This finding provides interesting clues about the forces that rule cell division and the ways in which proteins can accomplish a particular role. And for those who only understand science as a means of achieving immediate benefits for mankind, the practical application of the discovery is obvious: if we create a drug that specifically affects these novel proteins, we will eliminate all possibility for the parasite to reproduce itself into the host… without affecting at all the host’s proteins, since they are biochemically different. This is the most desired result for modern drug designers: finding differences at the molecular level, that affect processes which are basic for the life of the attacker. And because of the aforementioned common lineage for all organisms, it is a very rarely achieved target.
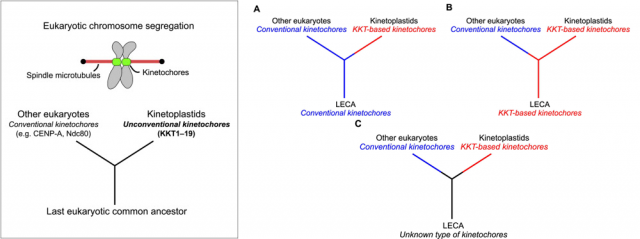
Last but not least, the finding is also extremely relevant regarding the evolution of the tree of life. It provides a beautiful example of how critical cell functions can be accomplished by molecular machineries that coincide in their final function, but differ in their composition; besides, it supports the idea that these organisms are older than the rest of eukaryotes. Although there is also the possibility that they simply lost the typical kinetochore proteins and further evolved towards possessing a different set of proteins to overcome this function, the authors of the work prefer the first option because it fits with other observations related with features of the mitochondrial complement of these organisms that are also quite different to the rest of eukaryotes. Are we then talking about the most ancient eukaryotic organisms still living in our planet? More research will be needed to finally get to that conclusion. In any case, this finding makes us a little closer to understanding how life on Earth evolved, which is always the most important lesson: it is impossible to understand ourselves without understanding how and from where we got to be like we are. Not to say that studying strong differences that still make us all part of a same huge family is an integrative lesson that benefits everyone, no matter if we are talking about your grumpy neighbor or the small parasite inside the mosquito eager to bite while you are sleeping.
References
- Rokas, A.; Carroll, S. B. Bushes in the tree of life. PLoS biology 4:e352; 2006. ↩
- Akiyoshi B. & Gull K. (2014). Discovery of Unconventional Kinetochores in Kinetoplastids, Cell, 156 (6) 1247-1258. DOI: 10.1016/j.cell.2014.01.049 ↩
4 comments
Mitosis without kinetochores? Wow! There are a lot of eukaryotic functions that seem rather complex, difficult to picture evolving from a prokariotic ancestor, and mitosis is one of them. Even if reduction cannot be rejected, I also like to see here such a complex structure evolving from simpler components. Like fancier and fancier chromosome-staples.
Thanks for your comment, pal!
I did not enter into detail, but looking carefully at the sequence alignments displayed in the supplementary material of the paper I also think that these proteins seem to be rather ancestral, since they are not so different to the rest of kinetochore proteins in other organisms. Surely there will be even weirder things out there, so don’t lose the faith in fiding a staple-like kinetochore someday!
[…] beheko aldean badaude oraindik aztertzen dabiltzan hutsuneak eta ondo ulertzen ez diren harremanak. Parasites, chromosomes and the tree of life-n Carlos Romá-Mateok ikerketa harrigarri bati heltzen dio, bizitzaren zuhaitzaren adar […]
[…] espacios en blanco y relaciones no del todo bien comprendidas que aún se están investigando. En Parasites, chromosomes and the tree of life Carlos Romá-Mateo aborda un estudio sorprendente que ayuda a conocer mejor una de las ramas más […]