Pet cafés and public health, new insights on bacterial resistance
Pet cafés and public health, new insights on bacterial resistance
Author: Diogo Roque is a PhD candidate at the Universidade de Lisboa.
Animal cafés are places where customers can enjoy food and drinks while interacting with animals such as cats, rabbits, or dogs. These cafés have become popular in many Asian cities, especially where apartments are small and the cost of living is high, making it difficult for young adults to own pets. In Europe, animal cafés are still not very common but have slowly started gaining attention, mainly through social media. Even though these cafés may look like just another cultural trend, they bring up an important global question: how do regular interactions between people and animals, particularly in public places, affect the spread of antibiotic-resistant bacteria? A recent study from Guangzhou, China, gives us some useful information and a warning about this issue 1.

In this study, the researchers studied three animal cafés in Guangzhou and took 111 samples from various sources: faeces and skin of animals and humans (customers and employees), as well as from surfaces like tables, bowls, and floors. From these samples, they identified 163 different bacterial strains. Escherichia coli was the most common species, found in 60 samples. While most E. coli strains usually live harmlessly in our intestines, some have virulence genes, making them capable of causing diseases, and antibiotic resistance genes, which means treatment becomes difficult. In this study, the researchers found something alarming: 90% of the E. coli strains were resistant to at least three different classes of antibiotics, making them multidrug-resistant. Furthermore, around 30% were classified as diarrhoeagenic E. coli, a group that is known to cause intestinal diseases.
Particularly worrying was the discovery of 19 E. coli isolates identified as belonging to sequence type ST328, which were all enteropathogenic (EPEC), meaning they can cause illness to humans. All these isolates came from one café and were found on human skin and faecal samples, and in animal droppings. Genetic sequencing showed these strains were differed only in a few single nucleotide polymorphisms (SNPs), suggesting recent transmission between humans and animals. Moreover, all these strains carried the virulence gene eae, which allows them to attach to human intestinal cells, confirming their potential to cause disease.
The study also explored how these bacteria carry and share antibiotic resistance. Many resistance genes were located on plasmids – small, circular pieces of DNA separate from the bacteria’s chromosomes. Plasmids can move easily between different bacteria in a process known as horizontal gene transfer. This ability helps antibiotic resistance spread quickly. Some of the identified plasmids were complex hybrids, probably created by combining genetic materials from different bacterial sources. A particularly concerning finding was a plasmid carrying the gene tet(X4), which makes bacteria resistant to tigecycline, an antibiotic usually reserved for very serious infections. Finding this gene in a city environment, far from rural or agricultural areas where it was previously reported, highlights how resistance genes can move into new places, carried by highly adaptable bacteria. Other genes found included blaCTX-M variants, known to resist third-generation cephalosporins commonly used in hospitals.
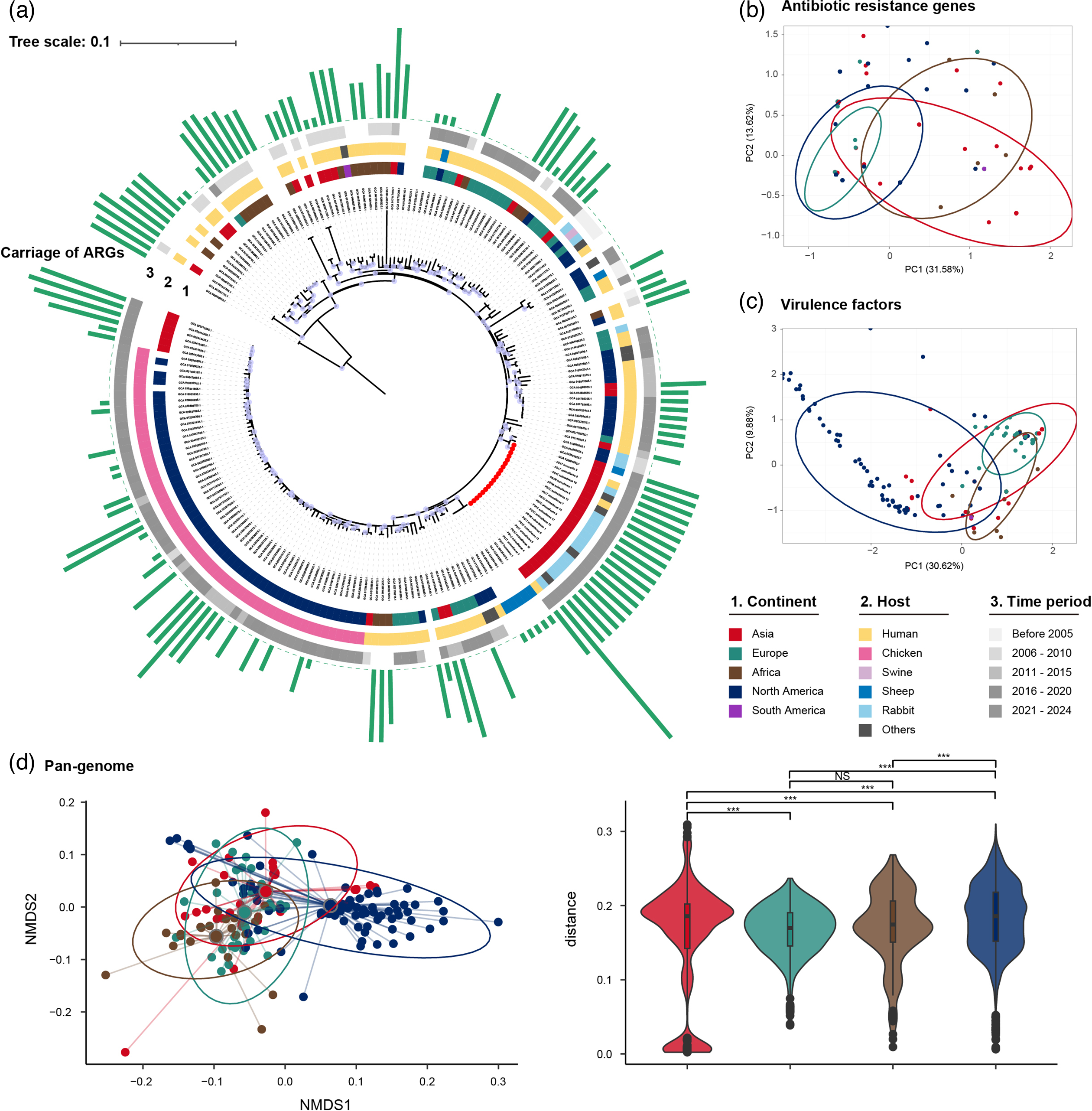
To better understand how widespread these bacteria are, the researchers compared their isolates to global databases of E. coli genomes (Figure 1). They found similar bacterial strains in livestock, wild animals, and human samples collected from many different countries. The similar resistance genes found in these international samples indicate that bacterial lineages like ST328 can spread widely across continents, adapting to various hosts and environments. The location of these resistance genes on easily movable plasmids likely helps this global spread.
The authors also pointed out several limitations of their study. First, they only examined three cafés, so their results might not apply everywhere. Second, they did not collect information about the cafés’ cleaning methods, the medical history or origin of the animals, or the behaviors of the visitors and staff. Without this information, it’s difficult to say exactly how the transmission of bacteria happens. Despite these limitations, finding nearly identical bacterial strains shared between animals and humans in public settings is significant enough to merit further investigation. The use of whole-genome sequencing helped the researchers detect transmission patterns that traditional culture methods could miss.
In conclusion, the researchers emphasized the importance of good hygiene practices in places where animals and humans come into close contact. This includes regular disinfection of shared surfaces, promoting hand hygiene among visitors, and potentially monitoring these venues for emerging antibiotic resistance. Although the study focuses specifically on animal cafés, the implications are much broader. It shows clearly that antimicrobial resistance can emerge not only in hospitals or farms but also in everyday urban settings.
Antibiotic resistance is a critical issue for global health, often spreading unnoticed until treatments become ineffective. Studies like this offer important insights into the small-scale interactions that allow resistance to persist and spread. They remind us that vigilance is necessary wherever humans and animals interact, not just in medical or agricultural environments. Whether or not animal cafés become common everywhere, the key message remains: bacteria do not respect human-made boundaries between species, locations, or ways of life.
References
- Sun, R.-Y. et al. (2025) The pet café is a neglected site for transmission of antimicrobial-resistant Escherichia coli in urban life. Microb Genom doi: 1099/mgen.0.001412 ↩