Regain the renewal capacity after myocardial ischemia? Pitx2 and its partners!
Author: Shu Ning got her BSc degree in Pharmacy from Shenyang Pharmaceutical University (China) in 2014. In 2016, she obtained her Master of Research degree on Clinical Research – Translational Medicine – at Imperial College London (UK) where she has worked on mitochondrial dysfunction in pulmonary smooth muscle cells from Pulmonary Arterial Hypertension. Currently she investigates on novel compounds and their pharmacological mechanism for treating cardiovascular diseases at the Pharmacy College, Tsinghua University (China).
Heart attacks cause millions of death annually around the world. Cardiac muscle cells (also named as cardiomyocytes) necrotized from myocardial infarction. Regenerating the dead cells, or to be more precisely, regenerative medicine is a newly flourished issue in recent two decades attempting to replace or regenerate cells, tissues or organs to restore its physical function. Some promising therapies has been developed in preclinical or clinical studies, such as cardiac progenitor cells and cardiac tissue engineering. Activating endogenous differentiating signalling can induce cardiomyocytes rejuvenation.
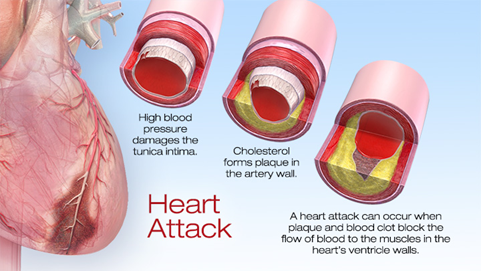
In a latest paper published in Nature by researchers from Baylor College of Medicine has reported a transcription factor Pitx2 that promotes cardiac repair1. Pitx2, paired-like homeodomain transcription factor 2, is known as one of the key mediators in developmental signalling; particularly Pitx2c, one of the three isoforms, regulates the asymmetric growth of heart, and its deficiency can cause the development of arrhythmia arterial fibrillation2. Then, how did they find out this transcription factor and what is the regulatory network involved? Let´s dig into this genetic pathway in heart regeneration.
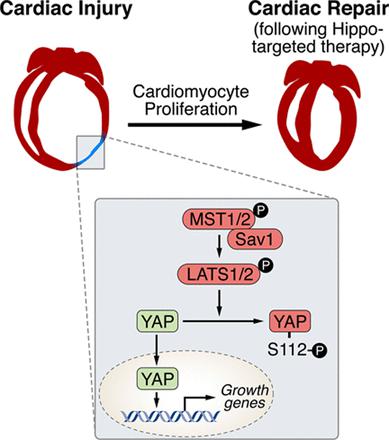
Ge Tao and his colleagues employed a regenerating heart model (Hippo-deficient hearts) as the starting point to find out the developmental transcription factors. What is Hippo-deficient regeneration model? Why they started from here? According to their published research early in 20133, Hippo signalling pathway has been reported as the endogenous repressor of adult cardiomyocytes regeneration. In that work, Salv, known as ‘Salvador’ (rescuer), a key component in Hippo signalling pathway, was mutated to verify the role of Hippo/Yap in heart development, regeneration and rejuvenation after myocardial injury 45. Based on that, authors continued to discover other regulatory molecules in this model. Throughout the work, LAD-O, short for left anterior descending artery occlusion, was performed to produce ischemia in postnatal mice heart.
To begin with, by using techniques such as immunofluorescence staining and RNA-sequencing, the researchers found enrichment of Pitx2 in the cardiomyocyte nuclei surrounding the infarcted area. From the available genomic data, Nrf2, a regulator of antioxidant response, was revealed enriched at the Pitx2 locus. After finding out these factors, two key questions emerged:
Question 1: is Pitx2 involved in the regenerating capacity of the injured heart?
Question 2: How does Nrf2 regulate Pitx2 after injury?
First, researchers verified that Nrf2 binds at the Pitx2 locus in the setting of ischemic hearts from mice after LAD-O. On top of that, they found a decrease in Pitx2 from both in vitro Nrf2 knockout P19 cell and in vivo Nrf2 loss-of-function mice thus confirming a direct regulation of Nrf2 to Pitx2.
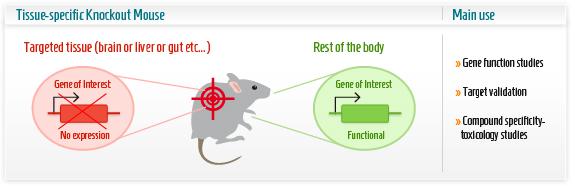
The research group then first concentrated on the renewal capacity of Pitx2 in neonatal heart regeneration, the ultimate event defined as the scaring and function of heart in this case. The research group employed a series of powerful techniques to create conditional knockout transgenic lines as animal model. In this part, understanding of the knockout techniques is critical. Cre-lox system consists of Cre recombinase that recombines a pair of short target sequences called Lox sequences. Floxing refers to the sandwiching of this Lox sequences to activate, represses the target gene in the transgenic animal model. This technique allows spatial and temporal alteration of gene expression in animal models and to link gene of interest to phenotypes.
Researchers used the MCKcre mice line, a muscle-specific knockout mice harbouring the Cre DNA recombinase gene. This Cardiac-specific tamoxifen-inducible Cre-expressing mouse lines has been developed as a conditional knockout transgenic mouse. In other words, by giving the mice an oral tamoxifen treatment, researchers were able to knock out the Pitx2 gene, specifically in the cardiac muscle, at the time after the infarct was conducted. These mice failed to regenerate.
However, it is well-known that mature cardiomyocytes have limited proliferative potential. So, researchers then continued to investigate whether the increased expression of Pitx2 was sufficient to restore the adult cardiomyocyte regeneration after ischemia injury. For this purpose, they created a Pitx2 overexpression model. A reduced scar, a complete morphology, a function recovery and an increased proliferation was demonstrated in the overexpressed model after ischemia compared to Pitx2 loss-of-function mice.
After investigating the regeneration capacity of Pitx2 in injured neonatal and adult heart, researchers tried to make an association of the Pitx2 and previously found factors Yap and Tead. Now it is known to us that Hippo signalling impedes adult heart regeneration (3). Early in 2013, investigators found heart regeneration when conditionally knocked out Salv in cardiomyotytes. Salv and Yap with its transfection co-activator Tead in the downstream Hippo signalling impact the cardiomyocyte proliferation and cardiac repair 67. Since similar capacity in neonatal heart regeneration of Yap from the previous work and Pitx2 in this study, authors wondered the exact cardiac renewal capacity of Pitx2 in this genetic-modified model. As expected, they found salv-Pitx2 double knockout heart failed to regenerate.
However, how is the network among these transcription factors, and is Yap also a binding partner to Pitx2? Co-occurrence of transcription factor binding motifs often suggests the interaction among transcription factors. Reading out from genomic data, motifs for Pitx2 and Tead have been found highly enriched and in close proximity and transcriptionally active, which was verified by purified glutathione S-transferase (GST) fusion proteins. That Pitx2 was a Yap binding partner was also confirmed from in vitro and in vivo aspects.
Hereto, the primary question concerning about the regeneration capacity of Pitx2 and its interaction with Yap, Tead in the injured heart has been answered. Then the second question comes: How does Nrf2 regulate the function of Pitx2 when triggering the cardiac regeneration after injury? First, they used P19 cells treated with H2O2 to mimic the state of increased oxidative stress in cardiomyocytes after ischemic injury. The data shows a translocation of Pitx2 from cytoplasm to nucleus, which depends on Nrf2.
Given the regulation from the upstream Nrf2 and transcriptional interaction from downstream Yap, we are eager to know whether there is any cross talk among Pitx2, Nrf2 and Yap. Researchers found that Nrf2-null hearts were unable to regenerate after myocardial ischemia, because antioxidant response is required. Nrf2-null hearts with Pitx2 overexpression failed to regenerate thus suggesting that Pitx2 promotes antioxidant response. Researchers also found that knocking down Yap expression compromised heart regeneration even when Pitx2 was overexpressed. Based on that, the connection between Nrf2, Pitx2 and Yap was clear: Nrf2 and Yap can regulate the Pitx2-mediated heart regeneration.
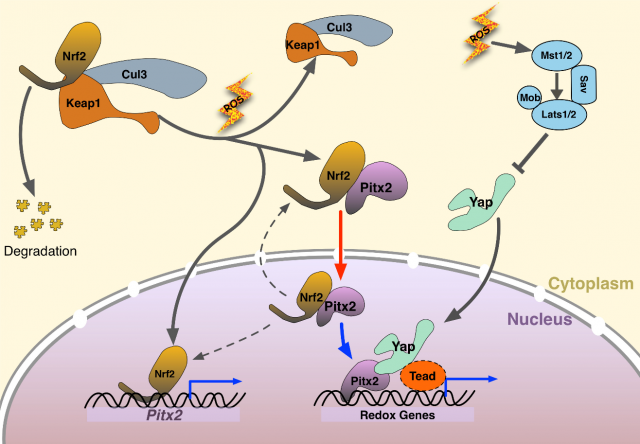
By now, it has been clarified about the renewal capacity of newly found Pitx2 and the interaction with its regulatory factors Nrf2 and Yap. What targets genes of Pitx2 are induced after injury that complete the heart regeneration? By RNA sequencing wild type and knock out hearts. The researchers found a set of oxidative stress response genes. A total of 505 direct target genes was reported to be activated in the downstream of Pitx2, mainly enriched in mitochondria, oxidation-reduction, and respiratory chain complexes, among which have redox scavengers such as superoxide dismutase genes Sod1 and Sod2 and glutathione peroxidase gene Gpx4. From the data of Yap ChIP-seq four redox genes were found to be bound by both Pitx2 and Yap. ChIP-seq (chromatin immunoprecipitation with DNA sequencing) is widely used to determine the transcription factors that affect the gene expression and the related phenotypes. This further confirmed a cooperative role of Pitx2 and Yap on activation of redox genes in the response to oxidative stress. Interpreting from the massive data, authors discovered high activity of ROS in apical border zones, which contributes to scarring in Pitx2-CKO hearts.
Finally, here we have the mechanism. When myocardial ischemia occurs, increased ROS is a natural oxidative response to ischemic damage. Nrf2 upregulates nuclear Pitx2 transcription and binds to cytoplasmic Pitx2, shuttling into nuclei. Pitx2 then recruits Yap to activate beneficial antioxidant scavenger genes, bestowing reparative capacity in myocardial ischemic damage. This paper revealed a significant genetic pathway for cardiac regeneration after myocardial ischemia injury. Reactivating the signalling pathway in the transient neonatal renewal process, regaining this regenerating capacity in mature heart indicates a potential therapy in cardiovascular disease. Whether this molecular finding on bench can be translated into clinical use? We shall keep an eye on it!
References
- Tao G, Kahr PC, Morikawa Y, Zhang M, Rahmani M, Heallen TR, et al. Pitx2 promotes heart repair by activating the antioxidant response after cardiac injury. Nature. 2016;534(7605):119-23. DOI: 10.1038/nature17959. PMID: 27251288 ↩
- Clauss S, Kaab S. Is Pitx2 growing up? Circulation Cardiovascular genetics. 2011;4(2):105-7. DOI: 10.1161/CIRCGENETICS.111.959791. PMID: 21505199 ↩
- Heallen T, Morikawa Y, Leach J, Tao G, Willerson JT, Johnson RL, et al. Hippo signaling impedes adult heart regeneration. Development. 2013;140(23):4683-90. DOI: 10.1242/dev.102798. PMID: 24255096 ↩
- Lin Z, Pu WT. Harnessing Hippo in the heart: Hippo/Yap signaling and applications to heart regeneration and rejuvenation. Stem cell research. 2014;13(3 Pt B):571-81. DOI: 10.1016/j.scr.2014.04.010. PMID: 24881775 ↩
- Heallen T, Zhang M, Wang J, Bonilla-Claudio M, Klysik E, Johnson RL, et al. Hippo pathway inhibits Wnt signaling to restrain cardiomyocyte proliferation and heart size. Science. 2011;332(6028):458-61. DOI: 10.1126/science.1199010. PMID: 21512031 ↩
- Papizan JB, Olson EN. Hippo in the path to heart repair. Circulation research. 2014;115(3):332-4. DOI: 10.1161/CIRCRESAHA.114.304389. PMID: 25035132 ↩
- Xin M, Kim Y, Sutherland LB, Murakami M, Qi X, McAnally J, et al. Hippo pathway effector Yap promotes cardiac regeneration. Proceedings of the National Academy of Sciences of the United States of America. 2013;110(34):13839-44. DOI: 10.1073/pnas.1313192110. PMID: 23918388 ↩
1 comment
[…] un texto muy interesante de Shu Ning sobre cómo recuperar los tejidos cardiacos tras un infarto: Regain the renewal capacity after myocardial ischemia? Pitx2 and its partners!. El Dr. Eduardo Oliver actuó de revisor del […]