How to model G-quadruplexes
How to model G-quadruplexes
Deoxyribonucleic acid (DNA) is the genetic material of most living organisms, being a major constituent of the chromosomes within the cell nucleus and playing a central role in the determination of hereditary characteristics by controlling protein synthesis in cells. DNA is, thus, a nucleic acid composed of two chains of nucleotides in which there is a sugar, deoxyribose, four bases: adenine (A), cytosine (C), guanine (G) and thymine (T). The two chains are wound round each other and linked together by hydrogen bonds between specific complementary bases to form a spiral ladder-shaped molecule, usually called a double helix.
Two nucleotides on opposite complementary DNA that are connected via hydrogen bonds are called a base pair. In the canonical Watson-Crick base pairing, A forms a base pair with T and guanine G forms one with cytosine C in DNA. In RNA, T is replaced by uracil (U). Importantly, pairing is the mechanism by which codons on messenger RNA molecules are recognized by anticodons on transfer RNA during protein translation. Some DNA- or RNA-binding enzymes can recognize specific base pairing patterns that identify particular regulatory regions of genes. Hydrogen bonding is the chemical mechanism that underlies the base-pairing rules described above, but contrary to popular belief, the hydrogen bonds do not stabilize the DNA significantly and stabilization is mainly due to stacking interactions.
A DNA secondary structure is the result of base pairing interactions within a single nucleic acid polymer or among several polymers. As we mentioned above, biological DNA mostly exists as fully base paired double helices. In a non-biological context, secondary structure is a vital consideration in the nucleic acid design of nucleic acid structures for DNA nanotechnology and DNA computing, since the pattern of base pairing ultimately determines the overall structure of the molecules.
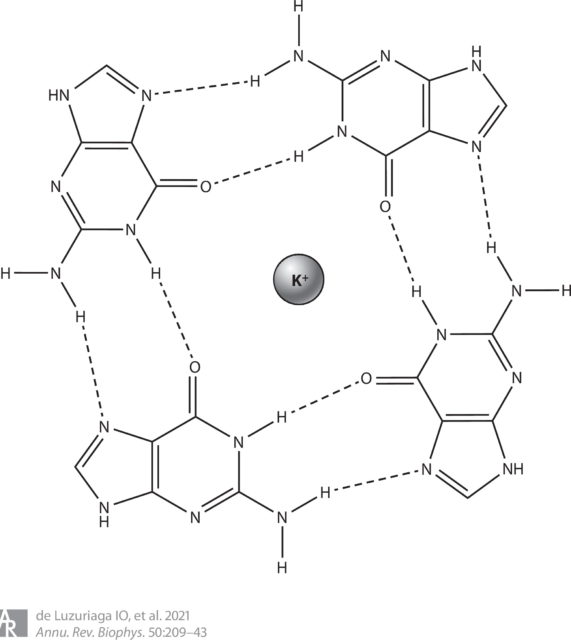
G-quadruplexes are secondary structures formed from nucleic acid sequences that are rich in guanine. The basic unit of these structures is known as a G-quartet or G-tetrad and consists of a square planar structure formed by four guanine bases, which interact via hydrogen bonds. Stacking of more than one G-tetrad forms the G-quadruplex. This stacking is produced in such a way that oxygen atoms of guanines are oriented to the centre of the structure, resulting in a tubular empty space, which is called the ion channel. When an alkali metal cation (typically potassium) is placed in this ion channel, it may interact with up to eight oxygen atoms; this coordination leads to the stabilization of the G-quadruplex.
To form the G-quadruplexes, the regular duplex DNA structure must open; once the helix is divided, the guanine-rich strand forms the G-quadruplexes. Although G-quadruplexes coming from a single strand are the most common structures, G-quadruplexes derived from up to four different strands have been found. G-quadruplexes have been associated with numerous genomic functions, such as transcription and replication, and studies have revealed their presence in key regions of the human genome, such as promoters, untranslated regions, and telomeres.
In the case of telomeres, the formation of telomeric G-quadruplexes has been shown to inhibit telomerase activity, which is responsible for maintaining the length of telomeres and is involved in 85% of cancers. This inhibition causes apoptosis of cancer cells and, since telomerase is overexpressed in the majority of cancer cells and in relatively few somatic cells, is recognized as a potential cancer-specific target that would avoid the death of somatic cells in chemotherapy treatments for tumours. For this reason, this strategy has been considered in current drug design to devise alternative therapies for cancer treatments, and the studies on small molecules that bind and stabilize G-quadruplexes have become a current hot topic of pharmaceutical and medical research.
To both carry out the inhibition of telomerase and alter the expression of oncogenes via a mechanism similar to that described above, G-quadruplexes have to be stabilized to ensure the disruption of these processes. Many organic small planar molecules and some metal complexes showed not only high affinity for G-quadruplexes, but also more selectivity for binding to G-quadruplexes than to duplex DNA, indicating that they may further stabilize the G-quadruplexes.
Different molecules, binding modes, and mechanisms of stabilization of G-quadruplexes have been studied during the past decades with the aim of understanding and rationalizing the formation and stabilization of these G-quadruplexes. In many of these investigations, experimental studies have been supported by computational methods for a better understanding of the formation of G-quadruplexes or their interaction with ligands.
Now, a team of researchers reviews 1 the state of the art of theoretical and computational methods, used to address these systems.
The researchers focus on in silico approaches and provide a broad compilation of different leading studies carried out to date by different computational methods. Both classical and quantum methods are considered. Classical methods allow for long-timescale molecular dynamics simulations and the corresponding analysis of dynamical information, whereas quantum methods (semiempirical, quantum mechanics/molecular mechanics, and density functional theory methods) allow for the explicit simulation of the electronic structure of the system but, in general, are not capable of being used in long-timescale molecular dynamics simulations and, therefore, give a more static picture of the relevant processes.
They conclude that computational methods are generally able to reproduce the experimental evidence and help provide a better understanding of the processes involving G-quadruplexes. But there is a caveat: we must be careful when selecting methods, force fields, or other parameters that affect the simulations and quantum studies, since they can lead to results that are not correct.
Author: César Tomé López is a science writer and the editor of Mapping Ignorance
Disclaimer: Parts of this article may be copied verbatim or almost verbatim from the referenced research paper.
References
- Ortiz de Luzuriaga I, Lopez X, Gil A. (2021) Learning to Model G-Quadruplexes: Current Methods and Perspectives. Annu Rev Biophys. doi: 10.1146/annurev-biophys-060320-091827 ↩