A mediator for horizontal gene transfer between eukaryotes and prokaryotes
A mediator for horizontal gene transfer between eukaryotes and prokaryotes
In biology few issues are as basic as the division and classification of living things. Despite the differences shown by taxa that occupy each of the categories, all living things have a common ancestor and characteristics that unite them.
Since Linnaeus (in 1735) to the newest division created by Cavalier-Smith (in the late twentieth century), they have tried to organize and classify living things. During this learning process, both fundamental concepts and standards (such as binomial nomenclature) were introduced not only for this classification but to the universal knowledge of biology. With all this knowledge about the species that inhabit our planet it was established that the domain would be the taxonomic category that would bring together the major groups of living things. This domain would be the upper echelon of the hierarchy detected in biological diversity.
At present, the more accepted proposal for scientists on the division of living things was introduced by Carl Woese in 1990, classifying them into three domains: Bacteria, Archaea and Eukarya, where Bacteria and Archaea belong to the ancient empire Prokarya (described by Édouard Chatton in 1937). Woese developed this classification based on the differences in 16S rRNA genes. Bacteria, Archaea and the eukaryotes each arose separately from an ancestor with poorly developed genetic machinery, often called as progenote. This biological classification is known as the three-domain system.
We find that living things have a fundamental feature that allow us to classify them into two large groups: prokaryotic (without nuclear membrane) and eukaryotic (including nuclear membrane).
These basic but huge differences make Escherichia coli, Thermoplasma acidophilum and Homo sapiens-for example- belong to different domains. A simple analysis of these organisms: their structure, habitat or type of nutrition -among other features- is enough to realize how different they are. All these data, scientifically proven, suggest that they are so different that it would be impossible to think that they have something in common. But it is not true.
They have a common link that has probably facilitated their evolution: a mediator. It has recently been brought to light a promising mediator (at least one of them) that makes possible horizontal gene transfer (HGT) between distant organisms like prokaryotes and eukaryotes.
The research 1, developed by Dr. Modesto Redrejo-Rodriguez from the group led by Margarita Salas and published in the Proceedings of the National Academy of Sciences (PNAS), shows that the terminal proteins (TPs) located in different groups of bacteriophage viruses (ɸ29, Nf, PRD1, Bam35 and Cp1) contain nuclear localization signals (NLS) involved in HGT between prokaryotic hosts of these viruses and eukaryotic organisms associated.
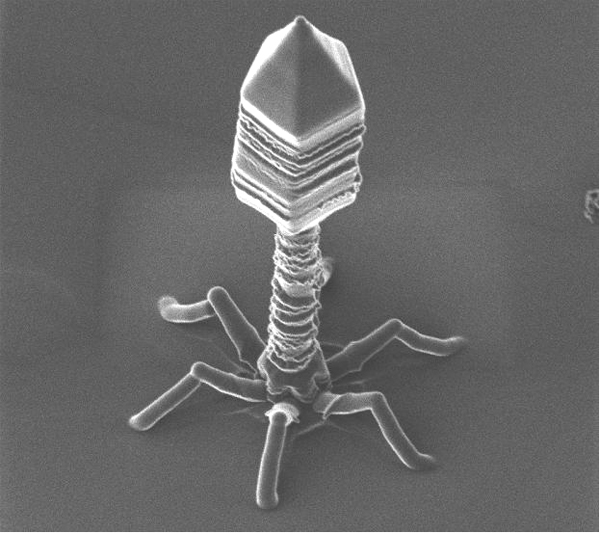
This transfer mechanism could have been important in the evolution of living things. Bacteriophage viruses -like any other viruses- are considered by many scientists as abiotic organisms. For that reason they are not included within the classification and division of living things. Bacteriophages are unusual because they infect bacteria and therefore have long served as a model in molecular biology for the study of gene transfer between different organisms.
The TPs of viruses (not only present in bacteriophage viruses) have different functions. E.g., Adenovirus (an icosahedral, nonenveloped virus family that contains a nucleocapsid, and a double-stranded linear DNA genome) have a TP involved in protein-primed DNA replication.
Phage ɸ29 is the main study model used by the group of Margarita Salas. This bacteriophage which infects the bacterium Bacillus subtilis has a TP (codified by gene 3) which is present in both ends of the phage DNA. This TP is used by the DNA polymerase of phage for the priming DNA replication, as in the case of adenoviruses.
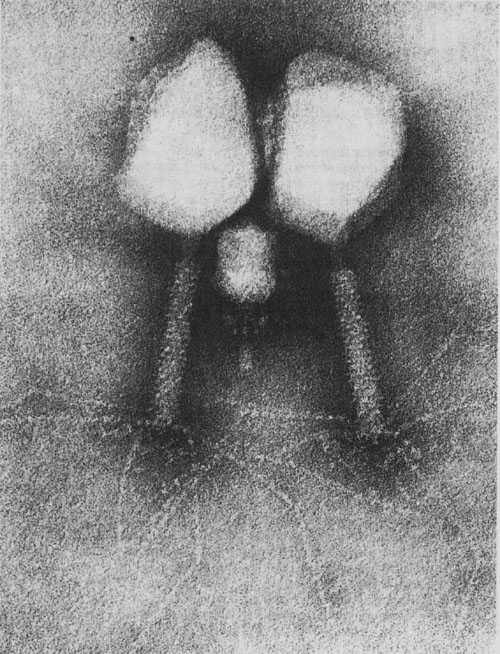
In addition, other non-bacteriophage viruses have a TP covalently attached to the 5 ‘ends of the nucleic acid whose function consists in the priming of viral replication in a similar manner to that produced in phage ɸ29. These viruses are -besides the human adenovirus mentioned above- poliovirus, encephalomyocarditis virus, Hepatitis B and C viruses and several groups of plant viruses. This indicates that the results obtained with phage ɸ29 could be extrapolated to these other non-bacteriophage virus families.
Likewise, the NLS are aminoacid sequences that serve to organisms to transport proteins to the nucleus through recognition of cellular proteins known as importins.
In some viruses which contain this NLS (e.g., lentiviruses), the ability to penetrate into the nucleus of host cells allow them to infect non-dividing cells. The capability to infect cells whose biochemical machinery is disabled gives these organisms an additional advantage, since most parasites, such as viruses, require exploiting the active biochemical machinery of cells they parasitize to complete its infectious cycle.
The presence of NLS in the genome of viruses that infect eukaryotic cells have biological sense. However this finding is non-expected in bacteriophage viruses due to they do not infect eukaryotic cells but prokaryotes; and these do not have a distinct nucleus. Therefore, this result -far from being expected by the researchers who conducted the study- raises new questions that could be related to the mechanisms of HGT between prokaryotes and eukaryotes.
In the work developed by Dr. Modesto Redrejo-Rodríguez et al., they generated 2YFP fusions to wild-type Φ29, Nf, GA-1, PRD1, Bam35, Cp-1, and ABV TPs in order to characterize the subcellular location when expressed in eukaryotic COS-7 cells. These experiments verified experimentally the nuclear localization of TP by confocal microscopy.
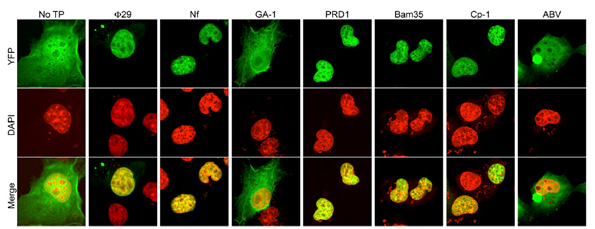
They not only demonstrated the NLS of phage TP was essential to nuclear location in eukaryotic cells but also they were able to map this nucleotide sequence. The researchers found that nucleotides 1-37 of the TP domain encoding the putative NSL were necessary and sufficient for nuclear localization of phage. This fragment is composed primarily of positively charged residues.
Therefore HGT between prokaryotes and eukaryotes mediated by phage ɸ29 TP may have had an enormous importance as fixing mechanism of evolutionary advantages in new lineages generated.
Since the functional mechanism of phage ɸ29 TP has been found to be similar to the mechanism in other viruses, this could have been a widespread and efficient mechanism of HGT for the evolutionary improvement of living things.
However, despite the interesting contributions that have yielded these results obtained using bacteriophage viruses as a model of study, it is necessary a further characterization of the true role of the NSL located in viral TPs, not only in bacteriophage viruses, as HGT agent between prokaryotes and eukaryotes in order to elucidate its evolutionary implications.
Finally, these results are not only important in the field of evolutionary biology. The use of the phage ɸ29 TP which includes the NSL could become a valuable tool for the transfer of nucleic acid fragments directly to the nucleus of eukaryotic cells. Moreover, the results described in the paper determined it would be enough yo use the NSL sequence including nucleotides 1-37 as a potential tool in biology. This biotool could have multiple scientific applications in the field of molecular biology and biotechnology, through transient protein expression, or in the biomedical field, with applications in gene therapy, among others.
References
- Redrejo-Rodríguez, M., Muñoz-Espín, D., Holguera, I., Mencía, M., Salas, M. “Functional eukaryotic nuclear localization signals are widespread in terminal proteins of bacteriophages”. PNAS. November 6, 2012 vol. 109 no. 45 18482-18487 ↩
6 comments
This is a very interesting finding from an evolutionary point of view; but as you mention, it may have critical implications as a future tool for research. Once again, nature provides us of valuable strategies which in many cases are far more effective that the ones we design ourselves. Anything capable of improving the low efficiency of DNA transfection into mammalian cells is for sure something to take into account.
Nice debut, boss! ;D
Hi Doc!
Thank you very much for your comment. It is very encouraging to see that this issue arouses some interest. As you say, these results are not only important from an evolutionary point of view but also biomedical. We’ll see what happens in the coming years.
A pleasure to share this kind of project with you!
Quique
Hi!!
I’m glad you found interesting my work. Very nice article, well-written and didactic. I just discovered this site and became addicted. Great!
Hi Modesto!
It is a pleasure to hear that things about the researcher who conducted the study. I am very glad about it. So, I would like to thank you for that kind of multidisciplinary works with such a high level.
Regards!
Quique
hi everyone!
Just to complete the information on (the idea we have about) gene delivery applications, we just published a short note about this topic: http://www.landesbioscience.com/journals/cib/article/22829/?show_full_text=true
Hi!
Thanks a lot, Modesto! It’s really interesting in order to complete the information of the article. I’ll take a look to write something at mapping ignorance.
Regards!
Quique