Not so junk DNA: microRNAs and schizophrenia
Not so junk DNA: microRNAs and schizophrenia
Back in 2001, the Human Genome Project gave us a complete readout of our DNA, unraveling that magic sequence of As, Gs, Cs, and Ts that contained all the information for making one of us. Thanks to that, we now know that in that sequence only 1.5% has information to make proteins, a fact that is quite astonishing since proteins are responsible for nearly every task of cellular life, including cellular shape, movement, ability to respond to the environment, maintenance of its DNA, regulation of cellular growth… they are basically responsible for everything a cell can do. So how is possible that this 1.5% of our genome has the information to do everything? What happens to the resting 98.5%? Is it useless?
The answer is – of course not! Although for many years this 98.5% has been called junk DNA 1, more and more we discover that it contains “functional elements” that are absolutely necessary for regulating the 1.5% that codes for proteins. This elements are so important that the need to understand them has led to the creation of an international project called ENCODE (the Encyclopedia Of DNA Elements), which tries to catalogue every letter within the genome that we think does something.
One example of a functional element of the non-coding DNA region is the microRNA. This small piece of RNA does not create a protein, but it is able to change the amount of one or several proteins, therefore changing the biology of a cell. How? Pretty simple: it is able to interfere with the process of protein generation. Briefly, proteins are generated in a 3-step process: 1) Information from DNA is copied into dispensable RNAs (pre-mRNAs), 2) those RNAs are edited (mRNAs) and 3) edited mRNAs are translated into proteins. In the case of the microRNAs, the process is similar but there is no step 3. Instead of that, the edited microRNA is able to bind directly an mRNA with a very similar sequence stopping it from going to step 3 and avoiding new protein synthesis. This is pretty exciting biology since we now know that interfering with endogenous microRNAs could be used in therapy and several pharmaceutical companies are now following these approaches for developing drugs for diseases such as atherosclerosis, heart failure, fibrosis or hypercholesterolemia 2.
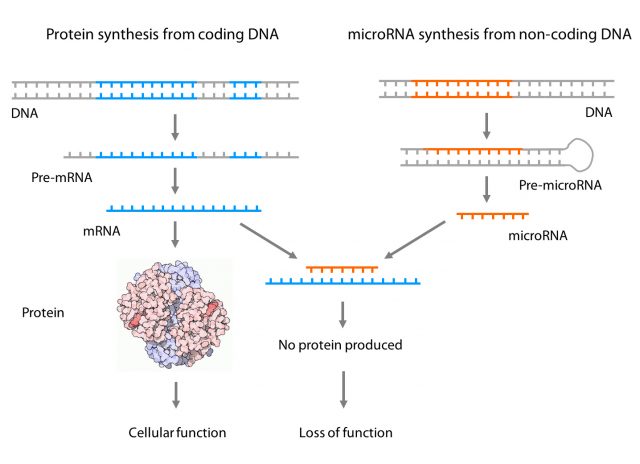
Therefore, understanding the role of microRNAs in health and disease is as important as understanding the role of proteins themselves. Dysregulations of certain microRNA functions are associated with the development of socially important diseases such as cancer, diabetes, Parkinson’s or Alzheimer’s disease and more and more we realize how much we still need to explore in this area. Even more, some diseases are genetically very complex (mutations in different genes result in the same disease) and microRNAs add a new layer of complexity since each microRNA can regulate, on average, the expression of approximately several hundred target genes 3.
One example of genetic complexity in disease is schizophrenia4. The current belief is that there are several genes that contribute to pathology or susceptibility of schizophrenia, but none exhibits full responsibility for the disease. Recently, after sequencing the entire genome of more than 20.000 schizophrenia patients, it has been found5 that the most common altered region in the DNA of these patients was a non-coding region (i.e. it was not a mutation in a gene that creates a protein). This region, which would have been considered junk DNA a few decades ago, turned out to have information to produce a microRNA named miRNA-137 that seems to contribute to schizophrenia susceptibility.
But how is miRNA-137 related to schizophrenia? How is it affecting the brain or neuronal function? This question was solved a few weeks ago when a group at the Massachusetts Institute of Technology (MIT) discovered that this microRNA regulates the presence of three key neuronal proteins: complexin-1 (Cplx1), Nsf and synaptotagmin-1 (Syt1). These three actors are absolutely essential for the communication between neurons since they modulate the release of neurotransmitters (the chemical signals that mediate neuronal communication). The researchers showed that when more miRNA-137 is present, neurons have fewer amounts of these three key regulators and their ability to transmit information to neighboring neurons is severely affected 6.
Schizophrenia has long been proposed to be a disease in which brain connectivity and neuronal communication is affected 7. We already knew that many key proteins are involved in regulating brain connectivity and neuronal function but now, thanks to these studies, we know that neuronal intercommunication can be regulated by microRNAs and that improper regulation of these small guys can severely affect brain function, increasing the susceptibility to suffer schizophrenia. So, I guess you clearly agree with me: Not so junk DNA, right?
References
- Palazzo, A. F., & Gregory, T. R. (2014). The case for junk DNA. PLoS genetics, 10(5), e1004351. doi:10.1371/journal.pgen.1004351 ↩
- van Rooij, E., Purcell, A. L., & Levin, A. A. (2012). Developing microRNA therapeutics. Circulation research, 110(3), 496-507. doi:10.1161/CIRCRESAHA.111.247916 ↩
- Meola, N., Gennarino, V. A., & Banfi, S. (2009). microRNAs and genetic diseases. Pathogenetics, 2(1), 7. doi:10.1186/1755-8417-2-7 ↩
- Ripke, S., O’Dushlaine, C., Chambert, K., Moran, J. L., Kähler, A. K., Akterin, S., … & Steinberg, S. (2013). Genome-wide association analysis identifies 13 new risk loci for schizophrenia. Nature genetics, 45(10), 1150-1159. ↩
- Schizophrenia Psychiatric Genome-Wide Association Study (GWAS) Consortium. (2011). Genome-wide association study identifies five new schizophrenia loci. Nature genetics, 43(10), 969-976. doi:10.1038/ng.940 ↩
- Siegert, S., Seo, J., Kwon, E. J., Rudenko, A., Cho, S., Wang, W., … & Tsai, L. H. (2015). The schizophrenia risk gene product miR-137 alters presynaptic plasticity. Nature neuroscience. doi:10.1038/nn.4023 ↩
- Frankle, W. G., Lerma, J., & Laruelle, M. (2003). The synaptic hypothesis of schizophrenia. Neuron, 39(2), 205-216. doi:10.1016/S0896-6273(03)00423-9 ↩