SNPs, RNA and Celiac Disease
Author:
Ainara Castellanos-Rubio obtained her phD on genetics of autoimmune diseases at the University of the Basque Country in 2010. Then she moved to NY where she did her postdoctoral training in the laboratory of Prof. Sankar Ghosh at Columbia University. Now she is back to the UPV-EHU as a beneficiary of a Juan de la Cierva incorporation fellowship in Dr. Jose Ramon Bilbao’s lab.
 Around 80% of the single nucleotide polymorphisms (SNPs) associated to many human diseases map to non-coding regions. Long non-coding RNAs (lncRNAs) represent a large portion of non-coding regions across the genome, however the link between the disease-associated SNPs on lncRNA expression or function, and the implications for disease, remain uncharacterized. Celiac disease is complex immune mediated disorder that develops in genetically susceptible individuals. This study1 describes for the first time the relationship between the function of a novel lncRNA that is present at very low levels in celiac patients and an associated polymorphism located within its sequence.
Around 80% of the single nucleotide polymorphisms (SNPs) associated to many human diseases map to non-coding regions. Long non-coding RNAs (lncRNAs) represent a large portion of non-coding regions across the genome, however the link between the disease-associated SNPs on lncRNA expression or function, and the implications for disease, remain uncharacterized. Celiac disease is complex immune mediated disorder that develops in genetically susceptible individuals. This study1 describes for the first time the relationship between the function of a novel lncRNA that is present at very low levels in celiac patients and an associated polymorphism located within its sequence.
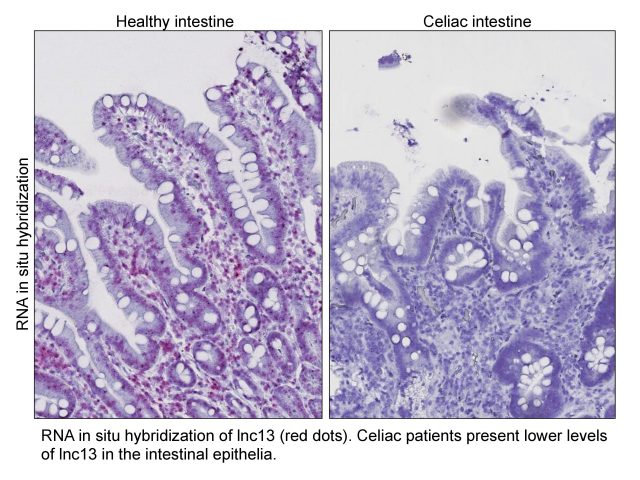
Genome Wide Association Studies (GWAS) have described around 40 regions associated with risk to develop celiac disease, several of which are located in noncoding regions of the genome2. SNP rs917997 is located 1.5 kb downstream of the gene IL18RAP, a gene that has been related to celiac disease and other autoimmune pathologies. Knowing that equivalent region in mice presents an actively transcribed lncRNA (lnc13), we analyzed its expression in human cells. The transcription of human led to the discovery of a novel non-coding gene harboring a celiac disease associated variant.
Although both genes overlap and are transcribed from the same DNA strand, transcription of lnc13 and IL18RAP are independent from each other. IL18RAP expression is induced in biopsies of celiac disease patients and in macrophages treated with an immune response stimulator. However, under the same conditions, the expression of lnc13 is reduced. The observed reduction of lnc13 is dependent on the transcription factor NFkB, which is known to be constitutively active in the intestinal epithelium of celiac patients (even on gluten-free diet) 3. Activated macrophages and celiac patients also showed an induction of the protein Dcp24, a negative regulator of RNA stability that is involved in the destabilization and consequent low levels of lnc13. In summary, we concluded that celiac patients present reduced lnc13 levels due to the destabilization mediated by Dcp2 and the constitutive activation of NFkB.
We also observed that lnc13 levels were negatively correlated with the levels of various inflammatory genes, some of them significantly overexpressed in biopsies of celiac patients. We could confirm that the function of lnc13 is to inhibit the expression of this subset of inflammatory genes in cells in which the immune response is not active. As has been observed with other known lncRNAs, lnc13 is mainly located in the cell nucleus and silences gene expression by binding to chromatin factors56. Specifically, lnc13 forms a complex with the protein hnRNPD (heterogeneous nuclear ribonucleoprotein D) and the enzyme histone deacetylase 1 (HDAC1). In non-stimulated cells, the repressive complex formed by lnc13 is localized to the promoters of the inflammatory genes, maintaining them transcriptionally inactive. In inflammatory conditions lnc13 is degraded, the complex is released and the inflammatory genes are expressed. This study shows how lnc13, hnRNPD and HDAC1 are bound to the promoters of certain genes but is still unclear if this lncRNA can target more genes and where the specificity of the binding lies.
The activity of this lncRNA is controlled by its interaction with the protein hnRNPD and this interaction we showed that this intereaction is dependent on the disease-associated SNP. In the presence of the risk allele rs917997*T, the binding of the lncRNA to the protein is weaker, which in turn would reduce its ability to regulate the expression of target genes increasing inflammation. This discovery underlines the importance of the three dimensional structure of RNA molecules, and suggests that small changes in RNA sequence can have an important functional impact.
In brief, this study shows how lnc13, a lncRNA that was previously unknown, is altered in celiac disease and depicts how a SNP that is associated with a complex disease can directly affect the function of a lncRNA. Other SNPs that have been associated with risk of developing common diseases are likely to function in a similar way but we cannot yet predict how this non-coding RNA molecules function. Further experimental studies on lncRNA function will help decipher the molecular principles of these RNAs and will allow the design of new therapeutic approaches.
References
- Castellanos-Rubio et al (2016) A long noncoding RNA associated with susceptibility to celiac disease Science doi: 10.1126/science.aad0467 ↩
- Trynka, G. et al. Dense genotyping identifies and localizes multiple common and rare variant association signals in celiac disease. Nat Genet 43, 1193-1201, doi:10.1038/ng.998 (2011). ↩
- Fernandez-Jimenez, N. et al. Coregulation and modulation of NFkappaB-related genes in celiac disease: uncovered aspects of gut mucosal inflammation. Hum Mol Genet 23, 1298-1310, doi:10.1093/hmg/ddt520 (2014). ↩
- Li, Y., Song, M. G. & Kiledjian, M. Transcript-specific decapping and regulated stability by the human Dcp2 decapping protein. Mol Cell Biol 28, 939-948, doi:10.1128/MCB.01727-07 (2008). ↩
- Huarte, M. et al. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell 142, 409-419, doi:10.1016/j.cell.2010.06.040 (2010). ↩
- Carpenter, S. et al. A long noncoding RNA mediates both activation and repression of immune response genes. Science 341, 789-792, doi:10.1126/science.1240925 (2013). ↩
1 comment
[…] Se ha descrito por primera vez la relación existente entre un ARN largo no codificante que está presente en los pacientes celíacos. Ainara Castellanos, que ha realizado la investigación, nos lo cuenta en SNPs, RNA and Celiac Disease. […]