Molecular Detectives: discovering new ion channels (I)

Doing research in molecular biology is not straightforward, because most of the experimental work is based in the study of molecules which cannot be seen by the unaided eye. Instead, we rely on indirect evidence given by methods which are specific for each area of research.
Let’s focus here as an illustrative example at someone studying ion channels. This individual is approached by a physician explaining he has found a patient suffering from episodic ataxia with continuous muscle movement and transitory seizures. The scientist suggests a mutation in a potassium channel leading to abnormal hyperexcitability could be the underlying cause for the observed symptoms. He even points to Kv1.1 potassium channel as the possible cause, because he knows it has been previously linked to episodic ataxia. The physician then collects blood from the patient after getting written consent and proceeds to sequence the gene encoding for the aforementioned ion channel (they know to which chromosome this gene is mapped, of course). Lucky enough, they confirm the gene from the patient has a single nucleotide change resulting in the modification of one amino acid located within one of the transmembrane segments of the protein (a domain of the ion channel embebbed within the membrane). The scientist then proceeds to clone the mutated gene onto a bacterial vector and then introduce it into a cell line which will express the ion channel to assess its functional properties. To do so, he will use a technique called patch-clamp which allows to record the electrical currents generated by the channel within the cell when a voltage command is applied. In addition, he will be comparing the current obtained to that from the Kv1.1 potassium channel not harbouring the mutation, the so called wild type version. After his set of experiments, they come to the conclusion that the gating properties (activation) from the mutant channel are impaired, because the amino acid change is affecting a critical region called the voltage sensor, thus making it unable to “sense” differences in voltage. Hence, the neurons controlling the patient´s muscles stay repolarized much longer than they should, provoking the seizures and muscle movements.
For somebody with no scientific knowledge, this example may seem quite impressive, but perhaps more interesting (and fascinating) would be to imagine how did the scientist who first identified Kv1.1 managed to do so. How previously unknown proteins are identified among twenty-something thousand genes comprising our entire genome? Well, unfortunately there is not a single answer to that. Instead, one can design different strategies depending on the protein meant to be found. In the coming two posts, I will try to explain two of them which seem particularly remarkable to me, because they require a balanced blend of cutting-edge technology and exceptional talent from the scientific team involved.
Back in 1997, the group of Professor David Julius in San Francisco published a Nature paper identifying the non-selective cation channel TRPV1 for the first time, previously known as the capsaicin receptor 1. It had been known for decades there was a conductance (an electrical current) in DRG sensory neurons (the ones involved in conveying pain-related information from our body) activated by capsaicin, the main pungent ingredient found in chilli peppers. The scientific literature recognized
this conductance as being a key player in transmitting information regarding tissue damage to the CNS and could, therefore represent a valuable therapeutic target to alleviate pain in humans. However, its identity was still mysterious (remember, at the time the Human Genome Project was yet an ongoing effort).
David Julius had the bright idea of using capsaicin as a stimulus to find the gene expressing the capsaicin receptor. But how to find a gene of interest from the entire genome? Because only DRG neurons seemed to respond to capsaicin application, his team collected DRG ganglions from rodents (newborn rat and adult mice) and isolated the whole set of RNA messengers from them. The rationale was that the entire set of RNAs would include few RNA copies encoding for the capsaicin receptor itself. They proceeded then to create a cDNA library consisting of a set of complementary DNA cloned into bacterial plasmids encoding for the genes which are expressed on DRG neurons. Obviously, those cloned cDNA molecules would lack introns, as these are not transcribed into messenger RNA. Subsequently, they inserted all these clones into bacteria and grew them, obtaining 2.4 millions of independent bacterial clones expressing all the cDNAs. The next step consisted on dividing the whole set of independent clones into 144 pools, in order to allow the screening to be manageable. Summarising, they had 144 Eppendorf’s each of them containing approximately 17000 cDNA clones.
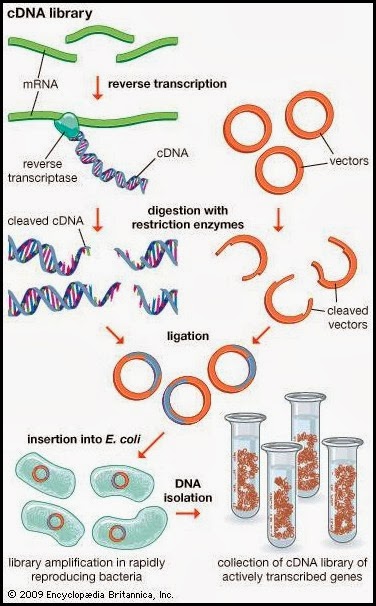
Perhaps the toughest part was to design a screening method to find the cDNA clone encoding for the capsaicin receptor, so here is when the talent came into play. They knew the receptor preferentially conducted Ca2+ ions under physiological conditions (they knew that form previous published studies) and they knew too about the existence of fluorescent dyes which once within a cell could produce fluorescence when bound to Ca2+. Therefore, if they could introduce the genes within a cell line unresponsive to capsaicin, along with the Ca2+-sensitive fluorescent molecule, they could then apply capsaicin and assess whether any of the cells let Ca2+ flow into the cell, thus producing a fluorescent signal after binding the fluorophore and indicating that the gene they were looking was there.
They chose a broadly used and well characterized cell line known as HEK-293 (human embryonic kidney cells) and injected the collection of cDNA clones contained within each pool into batches of them. The injected cells were plated on a glass coverslip and after inserting the fluorescent molecule into them, they recorded real time videos where at a specific time point, they applied capsaicin into the cells. If no cell within the image did show fluorescence it meant the clone was not present on the pool. However, when one or few cells showed a fluorescent signal right after applying capsaicin, it meant the cDNA encoding the capsaicin receptor was present on that particular pool (among the other 17000 clones). They would have then to pick that pool up and further subdivide it (thus, reducing the amount of clones per pool) repeating the experiment with every pool subdivision until they end up with one containing just a single cDNA encoding for the capsaicin receptor.
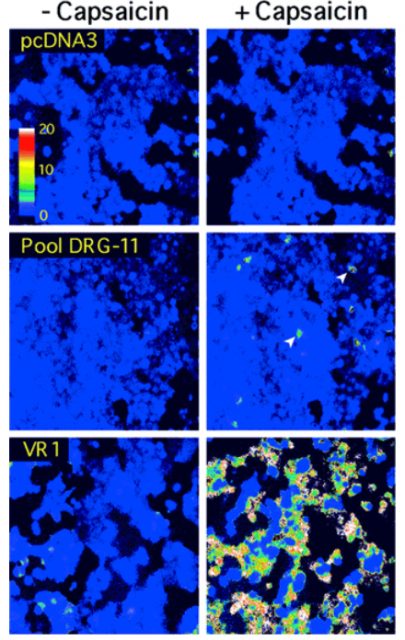
Their strategy was proven successful and after recording a substantial amount of videos, they ended up with a single 3-kilobase clone that, by itself conferred capsaicin sensitivity to HEK-293 cells. They named the cDNA VR1, for vanilloid receptor subtype 1, later known on as TRPV1. This discovery has had deep implications on the field of nociception (as well as others) and to date, around 4600 peer reviewed papers involving TRPV1 have been published, making an average of 242 per year. Unfortunately, we are still not able to completely and safely relieve patients from pain, but this is something we may have to deal with on another post.
References
- Caterina MJ, Schumacher MA, Tominaga M, Rosen TA, Levine JD, Julius D. (1997) The capsaicin receptor: a heat-activated ion channel in the pain pathway. Nature. 1997 doi: 10.1038/39807 ↩