Bacterial communities can store memories
Author: María Girbés Mínguez is a doctoral student at Center for Molecular Neurobiology Hamburg (ZMNH) / UKE (University Medical Center Hamburg-Eppendorf)
A recent study published in the journal Cell Systems 1 uncovered that bacterial communities (biofilms) can store complex patterns of information through membrane potentials at the cellular level, in a similar way to neurons. But before we dive into the study’s findings, let us remember some bacterial biology.
Biofilms and bacterial communication
Commonly, bacteria are represented as single cells that swim or float alone in a liquid medium, however, bacteria can behave as a community, and they often do so. Biofilms are formed once bacterial cells stick to one another, and while excreting the products of their metabolism, those become the extracellular matrix on which they establish and thrive, growing in huge numbers.

Bacteria in biofilms engage in distinct behaviours that were not evident in the past, when studies examined bacterial behaviour in low-density liquid cultures. The cells in bacterial communities show cooperative interactions that enhance collective survival 2. The structure of a biofilm as it is visible to the naked eye (figure 2) shows very intriguing features, like the way they grow into massive structured communities. In order to begin to understand how such organized communities are generated, we must examine the best characterized mechanism of bacterial communication: quorum sensing.
Quorum sensing (QS) aids bacteria in their transition to a community lifestyle. This system involves two components: a receptor in the surface of the bacteria capable of detecting the presence of a molecule, and a regulator of transcriptional response, which changes the amount or kind of proteins the bacterial cell produces for functioning 3. Since bacteria release molecules to the medium, these molecules can be detected by specific membrane receptors, allowing the bacteria to respond in specific ways to different molecules in the environment. QS is involved in many bacterial responses, such as antibiotic resistance, bioluminescence, or even the production of the necessary components to produce or maintain the molecular matrix of a biofilm4, therefore contributing to its appearance.
A key feature of quorum sensing is that the bacterial response is dependent of a threshold, so that when the extracellular concentration of a particular molecule reaches certain threshold is when the cells will respond. QS is therefore often promoting behavioural synchronicity in the population, so that a collective response is simultaneously generated in order to enhance virulence when the bacterial numbers are high, for example.
Conversely, bacterial communities can be tremendously heterogeneous, with cells present in different regions having different behaviours. A good example is the different behaviours of bacteria in the periphery of the biofilm, which have a different metabolic activity when compared to the cells of its interior, and these differences change over time as the biofilm develops and the cells cooperate in their metabolism.
Another important mechanism of bacterial communication is based on electrical signals, and it reminds us of the way neurons communicate. It is noteworthy that ion channels in bacteria and in mammalian cells are similar in structure. In fact the first characterized ion channel was the potassium ion channel from bacteria, detailed in the amazing paper by Rod MacKinnon and colleagues published in the journal Science 5. Although communication between neurons requires of both sodium and potassium ion channels, it is remarkable that potassium channels are commonly used for cell-to-cell communication in such distantly related cells.
The discovery of the function of potassium channels in bacteria is important to explain the differential behaviour of these cells depending on their position in the biofilm. Bacteria present in the interior of the biofilm are starving for glutamate, a vital molecule in bacterial metabolism which is less accessible for the cells packed in the interior6. When bacteria are in lack of glutamate, their potassium channels open releasing potassium to the exterior where the concentration is lower, and proton motor force facilitates the uptake by its specific transporter of glutamate together with two protons, aiding the cell3. The differences in membrane potential caused by the efflux of potassium, travel through the biofilm, where the efflux of potassium from one hyperpolarizing cell results in the influx of potassium by the neighbouring cell, depolarizing it, and this process results in electric oscillations.
Memory encoding in bacterial communities
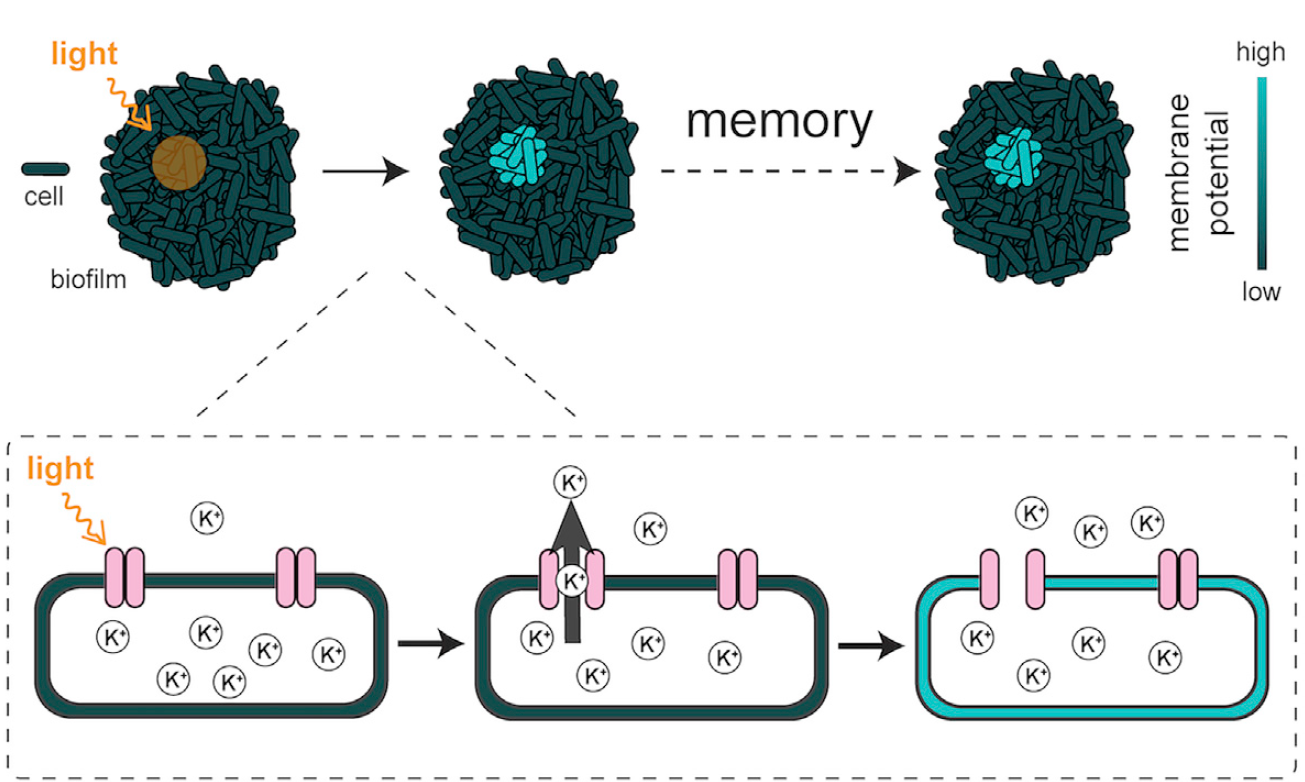
In this new study, the authors discovered that biofilms can store complex patterns of information. The researchers were aware that bacteria experience membrane potential spikes 7, and since other organisms have been found to experience changes in membrane potential upon exposure to blue light 8, they decided to test whether exposure to blue light could alter the membrane potential in biofilms of the bacteria Bacillus subtilis.
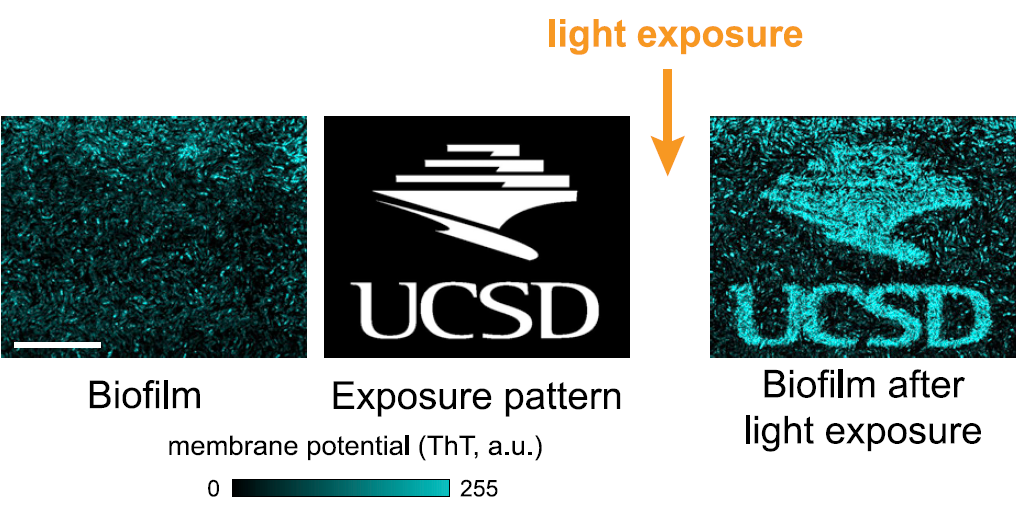
In order to test this, the authors studied a biofilm in conjunction with a special dye that reports differences in membrane potential by emitting fluorescence, and when areas of the biofilm were exposed to blue light these areas showed fluorescence indicating depolarization. Strikingly, the spatial pattern indicating a difference in membrane potential in the exposed area was persistent for periods of over 3 hours, implying that the biofilms can store patterns of spatial information in a membrane-potential-based memory, reminiscent of neurons. The biofilms can resolve fine details, as shown in figure 3.
The researchers also generated a mutant biofilm, lacking the only known potassium channel of B.subtilis, finding out that these cells did not show depolarization upon exposure to blue light, concluding that this potassium channel was responsible for the induced change in membrane-potential.
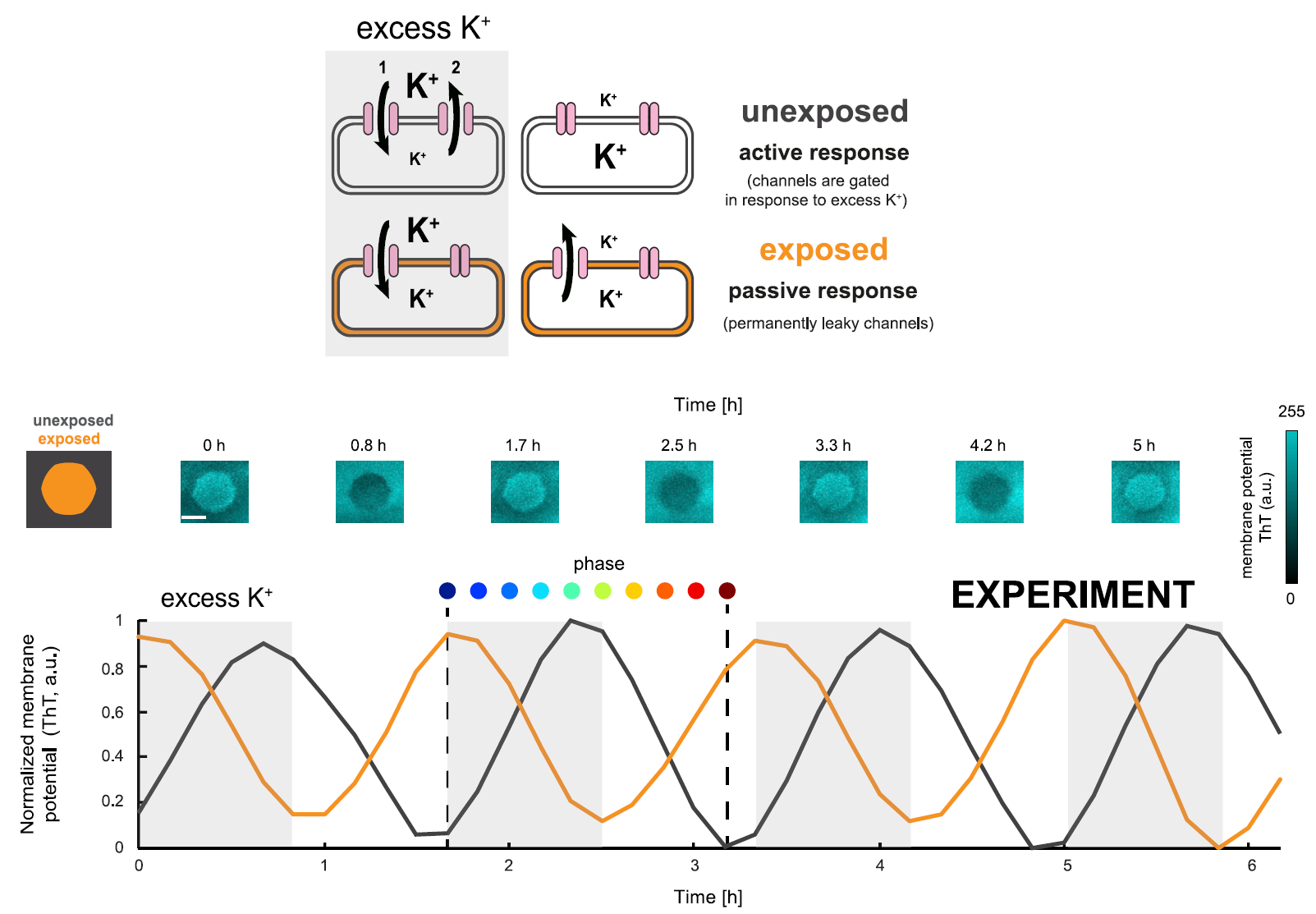
As explained previously, bacterial communities experience electric oscillations that travel through the biofilm as a mechanism of communication to alleviate starvation, responding to the individual requirements of glutamate, using potassium channels and varying the extracellular levels of potassium. An extraordinary finding is that the spatial memory encoded after light exposure in the biofilm is resistant to these electric oscillations. When responding to controlled changes in extracellular potassium concentrations, the cells that had been exposed to light showed a collective behaviour and switched their membrane potential in an opposite manner when compared to unexposed cells. Therefore, the information contained in the biofilm can be retrieved after hours, as seen in figure 4.
The findings of this study are valuable from an applied perspective, in the context of biological computing. Since optical perturbation of ion channels can be used to encode information in these bacterial cells, it may become a useful tool in order to achieve complex computations in bioengineering. Exposed cells respond in an opposite way to unexposed cells and can allow the encoding of ‘on’ and ‘off’ states as in traditional digital memory, so they could be used to coat synthetic circuits, performing computations via the use of light, or chemicals such as glutamine. This work contains promising findings that foresee a future in which we could use bacteria as memory-capable biological systems.
References
- Yang, C.-Y.; Bialecka-Fornal, M.; Weatherwax, C.; Larkin, J. W.; Prindle, A.; Liu, J.; Garcia-Ojalvo, J.; Süel, G. M. Encoding Membrane-Potential-Based Memory within a Microbial Community. Cell Systems 2020, S2405471220301162. doi: 10.1016/j.cels.2020.04.002. ↩
- Gloag, E. S.; Fabbri, S.; Wozniak, D. J.; Stoodley, P. Biofilm Mechanics: Implications in Infection and Survival. Biofilm 2020, 2, 100017. https://doi.org/10.1016/j.bioflm.2019.100017. ↩
- Gross, D.; Sevrin, A.; Shraiman, B. The Physics of Living Matter: Space, Time and Information in Biology. In The Physics of Living Matter: Space, Time and Information in Biology; WORLD SCIENTIFIC: Brussels, Belgium, 2020. ↩
- Berk, V.; Fong, J. C. N.; Dempsey, G. T.; Develioglu, O. N.; Zhuang, X.; Liphardt, J.; Yildiz, F. H.; Chu, S. Molecular Architecture and Assembly Principles of Vibrio Cholerae Biofilms. Science 2012, 337 (6091), 236–239. https://doi.org/10.1126/science.1222981. ↩
- MacKinnon, R.; Cohen, S. L.; Kuo, A.; Lee, A.; Chait, B. T. Structural Conservation in Prokaryotic and Eukaryotic Potassium Channels. Science, New Series 1998, 280 (5360), 106–109. ↩
- Bocci, F.; Suzuki, Y.; Lu, M.; Onuchic, J. N. Role of Metabolic Spatiotemporal Dynamics in Regulating Biofilm Colony Expansion. Proc Natl Acad Sci USA 2018, 115 (16), 4288–4293. https://doi.org/10.1073/pnas.1706920115. ↩
- Larkin, J. W.; Zhai, X.; Kikuchi, K.; Redford, S. E.; Prindle, A.; Liu, J.; Greenfield, S.; Walczak, A. M.; Garcia-Ojalvo, J.; Mugler, A.; Süel, G. M. Signal Percolation within a Bacterial Community. Cell Systems 2018, 7 (2), 137-145.e3. https://doi.org/10.1016/j.cels.2018.06.005. ↩
- Nagel, G.; Szellas, T.; Huhn, W.; Kateriya, S.; Adeishvili, N.; Berthold, P.; Ollig, D.; Hegemann, P.; Bamberg, E. Channelrhodopsin-2, a Directly Light-Gated Cation-Selective Membrane Channel. Proceedings of the National Academy of Sciences 2003, 100 (24), 13940–13945. https://doi.org/10.1073/pnas.1936192100. ↩
1 comment
[…] Eta bakterio-komunitateek memoria balute? María Girbések asalten du Bacterial communities can store memories […]