A closer look at peptide fibril assemblies
A new imaging technique can give scientists a much closer look at fibril assemblies, stacks of peptides like amyloid beta, most notably associated with Alzheimer’s disease.
These cross-β fibril assemblies are also useful building blocks within designer biomaterials for medical applications, but their resemblance to their amyloid beta cousins, whose tangles are a symptom of neurodegenerative disease, is concerning. Researchers want to learn how different sequences of these peptides are linked to their varying toxicity and function, for both naturally occurring peptides and their synthetic engineered cousins. Now, scientists can get a close enough look at fibril assemblies to see there are notable differences in how synthetic peptides stack compared with amyloid beta.
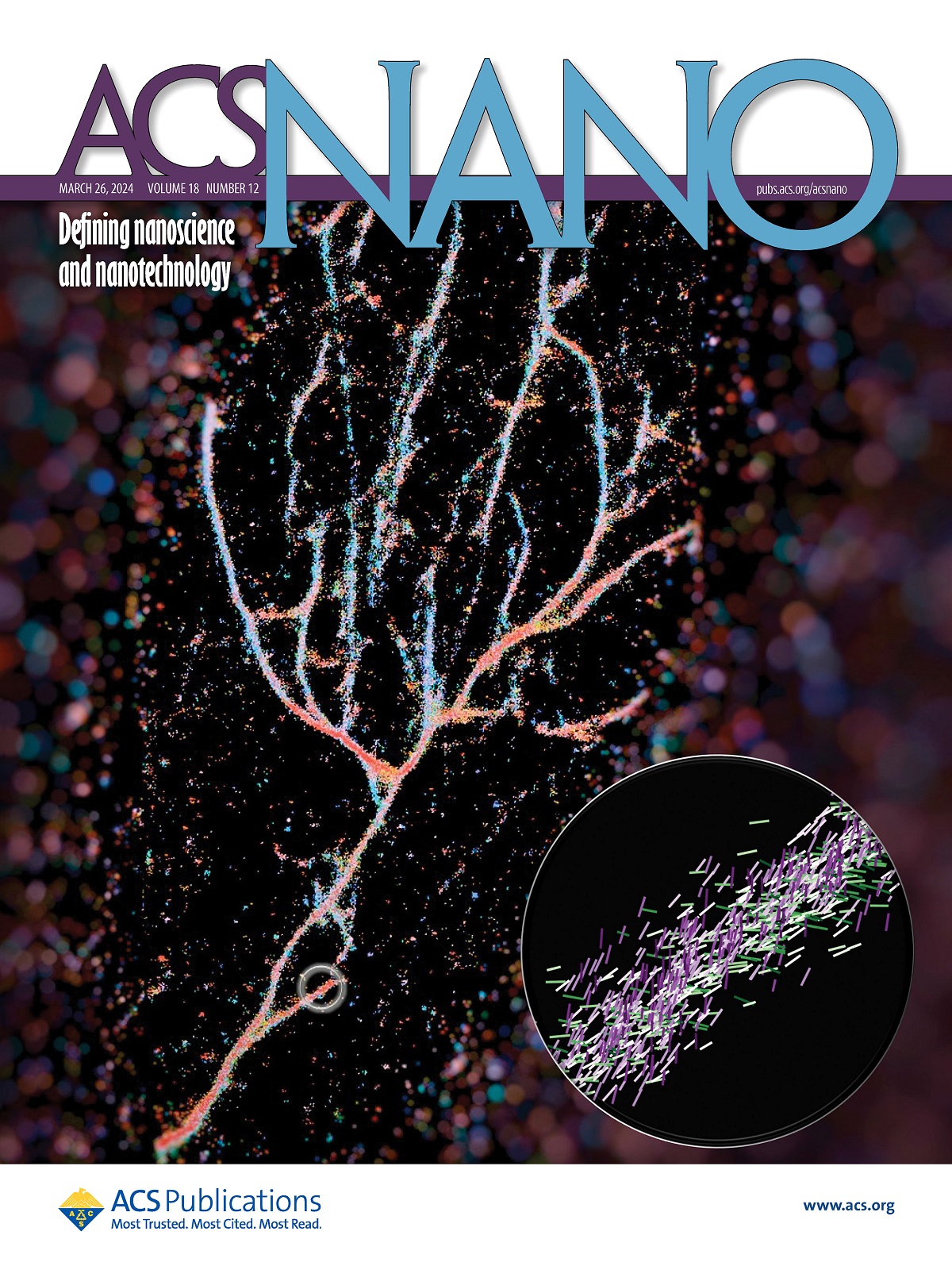
The researchers used the Nile red chemical probe to light up cross-β fibrils 1 . Their technique called single-molecule orientation–localization microscopy (SMOLM) uses the flashes of light from Nile red to visualize the fiber structures formed by synthetic peptides and by amyloid beta.
These assemblies are much more complicated and heterogenous than anticipated. But that’s good news, because it means there’s more than one way to safely stack your proteins. With better measurements and images of fibril assemblies, bioengineers can better understand the rules that dictate how protein grammar affects toxicity and biological function, leading to more effective and less toxic therapeutics.
First, scientists need to see the difference between them, something very challenging because of the tiny scale of these assemblies, impossible to discern using an optical microscope, or even some super-resolution microscopes.
A typical fluorescence microscope uses florescent molecules as light bulbs to highlight certain aspects of a biological target. In the case of this work, they used one of those probes, Nile red, as a sensor for what was around it. As Nile red randomly explores its environment and collides with the fibrils, it emits flashes of light that they can measure to determine where the fluorescent probe is and its orientation. From that data, they can piece together the full picture of engineered fibrils that stack very differently from the natural ones like amyloid beta.
References
- Weiyan Zhou, Conor L O’Neill, Tianben Ding, Oumeng Zhang, Jai S Rudra, and Matthew D Lew (2024) Resolving the Nanoscale Structure of β-Sheet Peptide Self-Assemblies Using Single-Molecule Orientation–Localization Microscopy ACS Nano doi: 10.1021/acsnano.3c11771 ↩