AINU, a powerful AI tool for studying cell heterogeneity
AINU, a powerful AI tool for studying cell heterogeneity
Chromatin is a complex of DNA and histone, a protein, in the nucleus of a cell. One of the main functions of chromatin is to help DNA packing. Cellular phenotypic heterogeneity is a key determinant of many biological functions; yet, it is still not clear whether it stems from the modifications of the chromatin structure or vice versa.
Cell heterogeneity can arise due to external agents that profoundly alter the chromatin structure of the host cells, such as viral infection – consider, for instance, that most of the world adult population is seropositive to herpes simplex virus type 1 (HSV-1). It also arises from chromatin alterations, which are a hallmark of cancer.
Thus, identifying cellular phenotypic heterogeneity can provide key information about biological functions, and revealing the chromatin structure of each cell can be a clear proxy of this heterogeneity.
Single-molecule localization microscopy (SMLM) methods, specifically stochastic optical reconstruction microscopy (STORM), has been used in the past to determine the nanoscale arrangements of chromatin fibres in cells Overall, SMLM represents a major advantage over diffraction-limited imaging, as it permits changes in nuclear nanostructures to be both visualized and quantified at the nanoscale. Current methods of analysing single-molecule spatial distribution, such as cluster algorithms, are powerful at extracting the nuclear locations and their local density. However, it is unclear how the spatial distributions and densities of these molecules can be used for identifying cell states.
Enter artificial intelligence. Convolutional neural networks have become the de facto standard for computer vision and a wide range of medical and healthcare imaging applications. Deep learning models have been used to classify whole-cell images and tracking using diffraction-limited microscopy. Actually, super-resolution microscopy has also been used to enhance localization precision during data acquisition and for semantic segmentation but not yet (to the best of our knowledge) to classify cells based on subcellular structures using SMLM images. Recently developed algorithms can now elucidate and interpret the outputs and decisions of deep learning models, enabling critical biological features to be identified and—importantly—overcoming the previous limitations of a lack of transparency and interpretability of results.
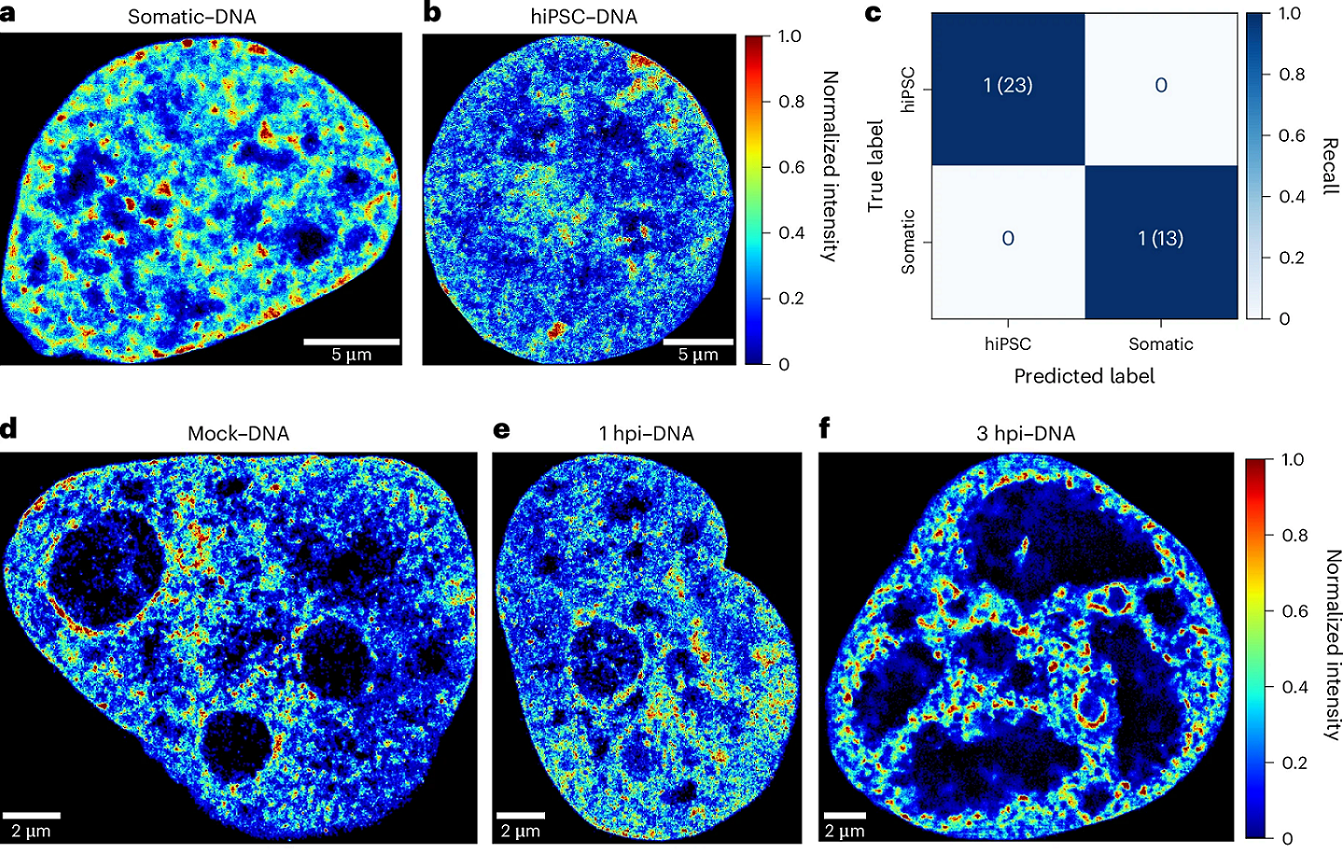
Now, a team of researchers takes 1 the next major step: they introduce the AI of the nucleus (AINU), a novel method that can effectively train a convolutional neural networks architecture using minimal training data from nuclear feature imaging.
The researchers employed STORM to image Ser 5-phosphorylated RNA Pol II, histone H3 and DNA in diverse cell states, including human somatic cells, human-induced pluripotent stem cells, human cells infected by HSV-1 and cancer cells. The approach combines convolutional neural networks with SMLM data to effectively identify cell heterogeneity and differentiate cell states.
Importantly, the model is interpretable. Models are interpretable when humans can readily understand the reasoning behind predictions and decisions made by the model. The more interpretable the models are, the easier it is for someone to comprehend and trust the model. Thus, this interpretable AI revealed that Pol II localizations within the nucleoli were the key feature recognized by AINU to identify human-induced pluripotent stem cells.
Overall, AINU emerges as a powerful tool for studying cell heterogeneity through SMLM nuclear imaging, demonstrating strong potential as a diagnostic method.
Author: César Tomé López is a science writer and the editor of Mapping Ignorance
Disclaimer: Parts of this article may have been copied verbatim or almost verbatim from the referenced research paper/s.
References
- Davide Carnevali, Limei Zhong, Esther González-Almela, Carlotta Viana, Mikhail Rotkevich, Aiping Wang, Daniel Franco-Barranco, Aitor Gonzalez-Marfil, Maria Victoria Neguembor, Alvaro Castells-Garcia, Ignacio Arganda-Carreras & Maria Pia Cosma (2024) A deep learning method that identifies cellular heterogeneity using nanoscale nuclear features Nature Machine Learning doi: 10.1038/s42256-024-00883-x ↩