O Phosphate! Where art thou?
O Phosphate! Where art thou?

Although we assume evolution of complex life forms as a matter of fact, we must admit that our brains are not ready for such an odd concept. Even though we can slightly imagine what a million year means, when we finally stare at some amazing life beings, we cannot fully comprehend the changes needed over an overwhelming amount of time to get to that final, beautiful animal or plant. The only way to try to understand the power of evolution is… to think smaller. It is impossible to imagine a complex structure appearing out of nowhere, but it is much easier to imagine a slightly simpler, previous one. If we follow this mental exercise, we might get a glimpse of how a very ancient, simple structure may evolve bit by bit, change by change, until its most complex and modern form.
The same exercise is useful when we talk about complex molecular structures and mechanisms: in particular, in the molecular strategy of protein phosphorylation. In multicellular eukaryotes, cells have developed an incredibly complex and ramified set of pathways in which molecules of phosphate are interchanged from on protein to the next one, acting as molecular switches, disruptors, bridges, and many other purposes. Phosphorylations – promoted by enzymes called kinases – affect very few amino acids among the twenty that form part of proteins. Especially common in mammalian organisms are phosphorylations in tyrosine, serine and threonine: these modifications control processes of all kind, driving protein interaction, degradation, or translocation to different cell compartments. These intricate phosphorylation cascades usually start with an extracellular stimulus that triggers the phosphorylation of a membrane protein, acting as a “sensor” of the particular conditions outside the cell. This sensor then phosphorylates a cytoplasmic counterpart inside the cell, and so on… It is difficult to imagine how such complicated and finely controlled processes may have evolved from previous, simpler components. But we’re going to make a big leap and observe what we can find in a very much simpler, smaller organism than ourselves: studying bacteria is sometimes like glancing through a window to the past, given that our ancestors were unicellular organisms.
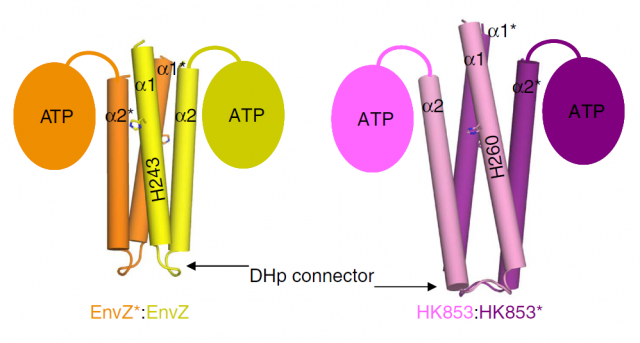
In bacteria, we can also find some phosphorylation events. Bacteria display a simpler form of “sensing” external stimuli, in which the phosphorylation they trigger is not in tyrosine, serine or threonine residues, but in histidines. Bacterial Histidine kinases (HKs) are dimeric proteins which respond to the external stimuli by phosphorylating themselves, in order to transfer the phosphoryl group to other protein (which is called response regulator, or RR) and thus promoting the cascade of events that will finally affect their genetic program: this is, in turn, what constitutes the response to the initial, outer stimulus. The structure of this kind of receptor proteins consists of two identical subunits with kinase activity, connected by a small loop called Dhp connector. So far, we only knew that some HKs start their signaling function phosphorylating the opposite subunit (this is then called trans-phosphorylation), whereas some others phosphorylated themselves (cis-phosphorylation). Since both subunits possess a catalytic core (the part of the structure where the phosphorylation reaction takes place), the fact that some proteins only performed cis reactions and others only trans, was striking. The group of Alberto Marina, at the Institute of Biomedicine of Valencia-CSIC, had already been pioneer in determining the molecular structure of a bacterial HK that they showed catalized a cis-autophosphorylation event 1. The information provided by this work was further exploited and just recently they publish a new structural study that finally unveils the mystery of cis and trans specificity in HKs. The results, published in Nature Communications 2, constitute a hard structural work with lots of chemical and physical details that may result overwhelming for the classical molecular biologist like the one writing these lines; fortunately, I’ve been lab-neighbor to Marina’s group for several years, so I could easily pass by and ask the main authors of the work, Patricia Casino and Laura Miguel-Romero, if they could kindly explain in easy words (i.e. for dummies) the strategy they used to unveil the mysteries of HKs. The result is the post you are reading right now.
Structural biologists pursue the crystallization of proteins, in order to determine the arrangement of their atoms in a three-dimensional landscape. This is commonly achieved by making an X-ray beam pass through those crystals and analyzing the scattering of that beam. Computers can do the final trick and calculate coordenates for the atoms that form the protein, making it possible to draw a model. This is like taking a snapshot of the protein, and after optimizing the process, one can try to obtain even more information by crystalizing the protein in complex with other things. In the case of proteins with kinase activity, one of the most interesting goals is obtaining the protein complexed with the phosphoryl group attached: thus, after studying the resulting snapshot, one can see which residues inside the protein are linking the phoshphate, and what other residues are chemically coordinated with it, finally being able to dissect the steps of the whole process. In fewer words: find the phosphate, and you will find the catalytic mechanism.
Our brave researchers had already described the structure of a HK from the bacteria Thermotoga maritime named HK853, and found that the phosphorylation took place in a cis-direction. Since there are other, very similar HKs that phosphorylate in trans-direction, they wondered whether both processes could be conditioned by different structural constraints of the particular proteins. They decided to use a very common strategy in molecular biology: they constructed a chimeric protein. It sounds more spectacular than it really is: it is very easy to manipulate genes to make combinations, so they easily fused the larger subunits of EnvZ, a very similar HK but which specifically phosphorylates in trans-direction, with the small connector of their favourite cis-HK, HK853. Making this hybrid protein, they just had to take the “structural snapshot” and observe if the HK kept working in a trans-direction as it always did, or the particularities of the “foreign” connector transformed the whole mechanism into a cis-phosphorylating event.
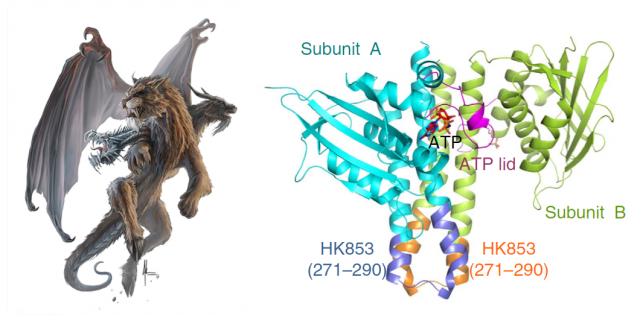
And interestingly, what they found was the latter. But what’s more, even though the EnvZ subunit of the protein had transferred the phosphate to itself, the linking between the phosphoryl group and the catalytic amino acids of the protein was identical to the one found in the natural EnvZ protein. A series of mutagenesis experiments followed by assays of enzymatic kinase activity served to compare the biochemical features of the native proteins with different chimeras, and to assess the function of each amino acid pointed out by the structural snapshots. In summary, the conclusion is that the catalytic mechanism is the same, and only the structure of the small loop connecting the subunits determines the direction of the phosphorylation, making it “fall” unto the same phosphorylating subunit or to the neighboring one, by a mere physical matter. The biological implications of this observation are very remarkable. We are talking about one of the most ancient sensory mechanisms in the unicellular world, and we observe that a slight set of changes in a small portion of a protein can drastically convert the result of a catalytic reaction. Since cis- and trans-phosphorylating HKs respond to different external stimuli in a specific manner (EnvZ responds to changes in the osmotic conditions outside the cell; however, the stimulus detected by HK853 remains unknown, despite all their similarities), the evolutionary potential becomes evident.
This is one of many beautiful examples of how small steps give rise to huge leaps. A slight substitution in a single nucleotide can change the resulting amino acid in a protein, and given the big differences in chemical properties between amino acids, this single change can make a part of a protein stiffen, bend, or simply gain or lose the ability to be phosphorylated. In a similar way, once the signaling pathways of HKs appeared, slight changes affecting the rigidity or size of Dhp connectors might have made it possible to respond to different stimuli, giving the bacteria bearing those changes new capacities to face the harsh outside world. Step by step, these molecular machines acquired complexity and suddenly became something much more suitable for regulation than before. Phosphate is one of the most abundant elements that form part of organisms. It is a component of the extremely stable structure of DNA, but also acts as a small molecule, acting like a signal, a message that passes from one protein to other, marking the path for other proteins. Following the phosphates inside a cell may represent a lot of answers to many biological questions. An odyssey we have just very recently begun, along which many wonders still lay unfold.
References
- Casino P, Rubio V, Marina A. Structural insight into partner specificity and phosphoryl transfer in two-component signal transduction. Cell. 2009 Oct 16;139(2):325-36. doi: 10.1016/j.cell.2009.08.032. Epub 2009 Oct 1. PubMed PMID: 19800110 ↩
- Casino P. & Alberto Marina (2014). Visualizing autophosphorylation in histidine kinases, Nature Communications, 5 DOI: http://dx.doi.org/10.1038/ncomms4258 ↩
2 comments
[…] Aunque asumimos que la evolución de formas de vida complejas es un hecho, debemos admitir que nuestros prejuicios cognitivos nos dificultan aceptar un concepto así con facilidad. La única forma de intentar comprender el proceso es…pensar en pequeño. Esto […]
[…] modu bakarra da… txikian pentsatzea. Hau da, Shakespearen imajinazio berbera erabiliz, O Phosphate! Where art thou? artikuluan iradokitzen diguna Carlos Romá-Mateok adibide zehatz bat proposatzerakoan: proteinen […]