The proper use of deep learning in microscopy image analysis
The proper use of deep learning in microscopy image analysis
Microscopy is a leading technology in biological research. Today, a typical microscopy session may generate hundreds to thousands of images, generally requiring computational analysis to extract meaningful results. But a simple analysis is not enough any more. Over the last few years, deep learning (DL) has increasingly become one of the gold standards for high-performance microscopy image analysis. DL algorithms have become powerful tools for analyzing, restoring and transforming bioimaging data. One promise of DL is parameter-free one-click image analysis with expert-level performance in a fraction of the time previously required. Is it a realistic promise? As with most emerging technologies, the potential for inappropriate use is raising concerns among the research community, what should we do to make sure the results provided are reliable and reproducible?
For image analysis, DL usually uses algorithms called artificial neural networks (ANNs). Unlike classical algorithms, before an ANN is used, it first needs to be trained. During training, the ANN is presented with a range of data, from which it attempts to learn how to perform a specific task. More specifically, the ANN builds a model of the mathematical transformation that needs to be applied to data to obtain the desired output. ANNs can therefore be considered as non-linear transformation machines, performing sequential mathematical operations on the input data. As we inspect deeper into these sequences of operations, it becomes difficult to understand what features of the original images are used. For that reason, they are often thought of as ‘black boxes’ as, for most users, only the input images and output predictions are readily available.
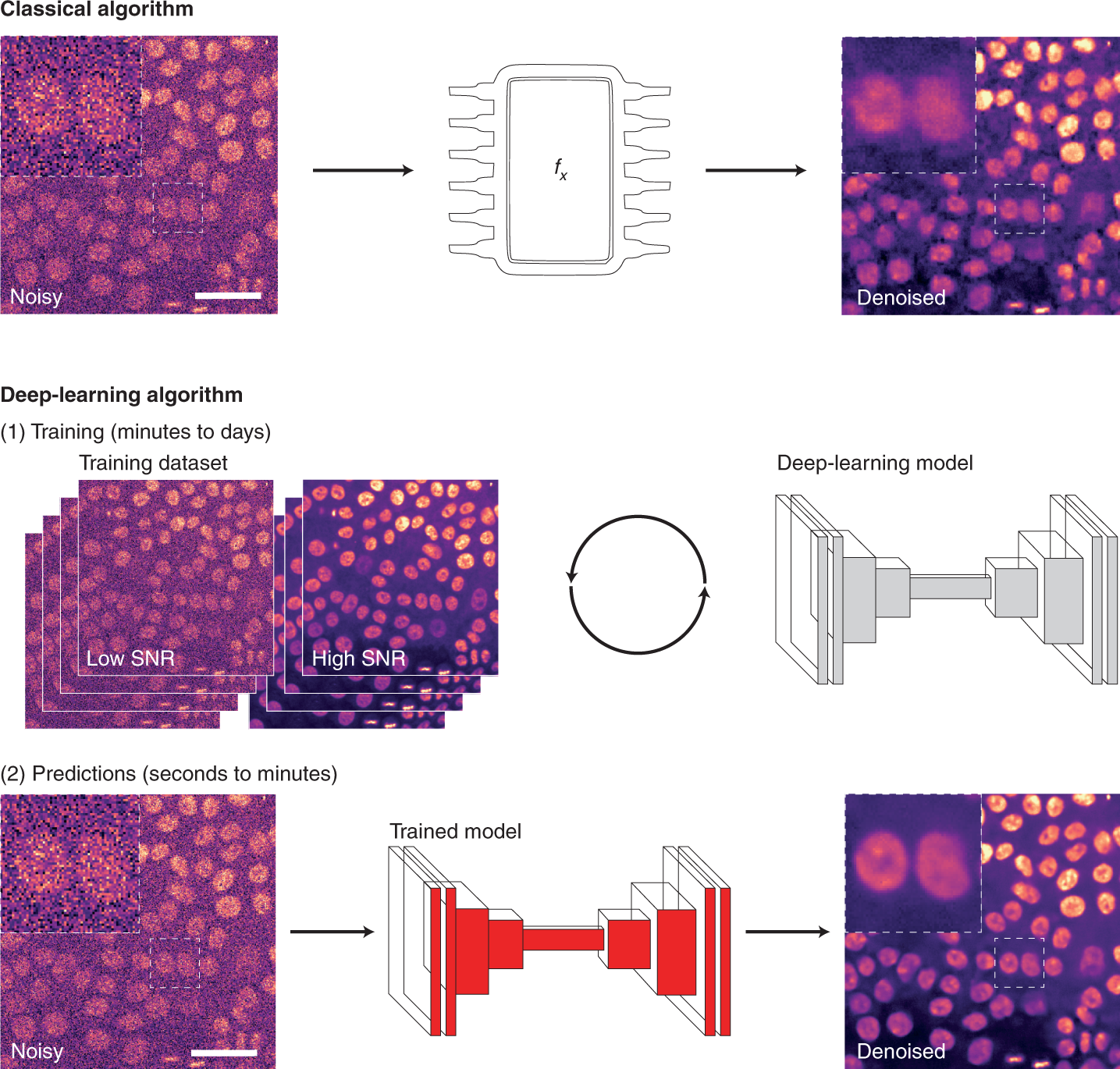
Learning how to perform an analysis from example data is both the principal strength and the main weakness of DL. By learning directly from the data, the ANN tries to identify the most suitable way to perform the analysis, leading to models with excellent performances for that particular dataset. However, trained DL models are only as good as the data, and the parameters used to train them. Thus, one powerful approach is to produce general models with high reusability potential using a large and diverse training dataset.
As DL models are becoming accessible through public repositories (so-called model zoos, such as bioimage.io) or web interfaces, it becomes straightforward to use the models directly to analyze new data. This has the advantages of speeding up DL uptake but, unless the researchers can confirm that their own data were well-represented within the training dataset used initially (which can be very difficult to do), the performance of such portable models on the new data often remains unclear.
This is the reason why despite its incredible potential, the application of DL in microscopy analysis has raised concerns due to a lack of transparency and understanding of its limitations, especially for generalizability. In addition to this, DL is developing at an incredible rate, which places a significant burden on users to determine the most appropriate tools for their needs. It remains challenging to assess the validity and performance of a range of approaches that are often difficult to compare, especially when widely accepted benchmark datasets are unavailable.
Now a team of researchers, in order to help define adequate guidelines and ensure the appropriate use of this transformative technology, has reviewed 1 key concepts that they believe are important for researchers to consider when using DL for their microscopy studies. They also describe in this work how results obtained using DL can be validated and propose what should, in their view, be considered when choosing a suitable tool. Finally, they also suggest what aspects of a deep learning analysis should be reported in publications to ensure reproducibility.
The researchers argue the importance of validating any model using a purposefully built evaluation dataset containing ground-truth target images or labels. Similarly, the use of DL models should be reported appropriately to ensure reproducibility and transparency. This is a challenging task for DL as many components, both internal (hyperparameters) and external (training dataset) to the network used, can dramatically influence the results obtained.
With the increasing availability of networks and models, finding ways to identify what might be a ‘good tool’ becomes critical. The authors believe that a good tool should not be only a performant one, but that its transparency of what it does to the data, usability and reliability are equally important.
The responsibility of proper use of DL in microscopy is now equally shared between users and developers. Spiderman’s Uncle Ben has never been more right than today: “With great powers comes great responsibility”.
Author: César Tomé López is a science writer and the editor of Mapping Ignorance
Disclaimer: Parts of this article may be copied verbatim or almost verbatim from the referenced research paper/s.
References
- Laine, R.F., Arganda-Carreras, I., Henriques, R. & jacquemet, G. (2021) Avoiding a replication crisis in deep-learning-based bioimage analysis. Nat Methods 18, 1136–1144 (2021). doi: 10.1038/s41592-021-01284-3 ↩