The benefits of recycling: neuron plasticity promoted by histone turnover
The benefits of recycling: neuron plasticity promoted by histone turnover
If there should be one rule that could apply to all fields of biological knowledge, it could be that they get more and more complex as we deepen into their study. Regarding genetics, the landscape has become not only more complex but also much more dynamic than initially thought. The old-fashioned concept of genes as reservoirs for information, which were kept secured and locked inside the cell nucleus, only to be used carefully for the production of proteins and to be passed on to the next generation, has been replaced by a dynamic view of structural rearrangements, time and space regulation, continuous copy and editing of the genome components and regulators. Genetic information is kept secured and locked, yes, but it is continuously being opened and closed again, used and re-used, and the number of editors, regulators and watchers has risen until providing a crowded environment, boiling with activity, that results in the amazing plasticity of the archetypal composition of any living cell. From this point of view, one of the cell types that has been found increasingly complex, variable and plastic is the neuron, and regarding nuclear components, the ones that are becoming more and more dynamic and surprise-bearers, are histones.
These small proteins were initially thought to have a merely structural role, holding together the DNA double helices in order to facilitate their condensation into a larger macrostructure called chromatin; but very soon, it was shown that this chromatin was not only necessary to re-arrange and condense the huge DNA molecules into an easy-to-handle travelling device, the chromosome – in order to perform adequate and even cell divisions. Chromatin packing and unpacking proved to be critical for the organization of genetic information and the manner in which proteins with a critical role in activating or inhibiting gene expression could interact with their target sequences. In the latest years, histone modifications and the so-called histone code, have become the focus of many research lines which provide a novel landscape of how specific and interconnected posttranslational modifications on the surface of histones modify their union to DNA. This strongly affects genetic expression and provides a critical level of regulation that modifies the cellular fate, in what has become one of the main branches on the field of epigenetics. Now, we step forward to find out that the capabilities of histones to modify gene expression can be critical in neurons, in a way that goes beyond the catalogue of usual and expected modifications.
As a reminder, we should mention that histones are bound to DNA in the form of octameric complexes constituted by different proteins; there is a “regular” set of histones, among which the one starring the work by Maze and collaborators is histone H3. Besides all the possible modifications on histone H3, an extra factor of variability is the existence of H3 variants named H3.1, H3.2, and H3.3 (which slightly differ in their amino acid composition). In this extensive work just published in Neuron 1, researchers have found the physiological relevance of these histone variants in brain plasticity. Since this cell type is characterized by not having an active cell cycle that requires unpacking, duplication and re-packing of their chromosomes (unluckily neuron populations do not regenerate with the facility of other cell types in our body), the possibilities of a specific histone-type turnover might hold the key to the already known capability of neuronal cells to create functional networks that result in the outstanding brain plasticity in vertebrates, during and after development. This was the starting hypothesis, and the results showed a robust and amazing confirmation.
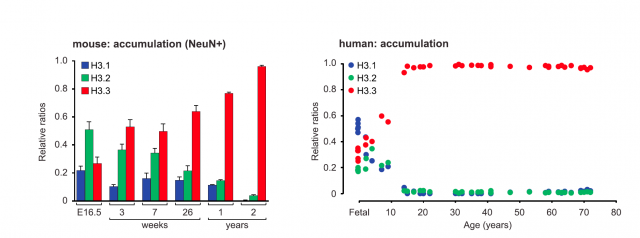
The work starts with a fine description of how the relative proportion between H3 forms varies with age, finding an increasing accumulation of the H3.3 variant both in neuronal cells from mice and human samples. This means that neuron chromatin in adulthood is enriched with histone H3.3, but besides, the pool of histone incorporated into chromatin is dynamic: old histone proteins are removed from chromatin and substituted by newly synthesized ones. And more interesting: whereas the abundance of histone versions H3.1 and H3.2 was mostly related to intergenic regions, H3.3 was found to enrich gene bodies and promoters of active genes. This was the first clue pointing to a functional role, beyond structural purposes, for histone H3.3 variant. To confirm this relationship, the researchers studied the active transcriptional regulation of H3.3 in response to neuronal stimulation. What they found was striking: quantitative PCR analysis showed a significant increase in the expression of the genes encoding H3.3 (there exist two genes that encode slightly different transcripts for H3.3) as compared to those encoding H3.1 or H3.2. And not only in cultured neurons but also in live mice subjected to chronic periods of environmental enrichment (a series of tests that promote the enhancement of synaptic plasticity and memory). These mice showed an increase in the incorporation of newly synthesized H3.3, specifically into their hippocampal region. These experiments pointed to a correlation between the exchange and recycling of histone types into the nucleosomes, and neuronal activity.
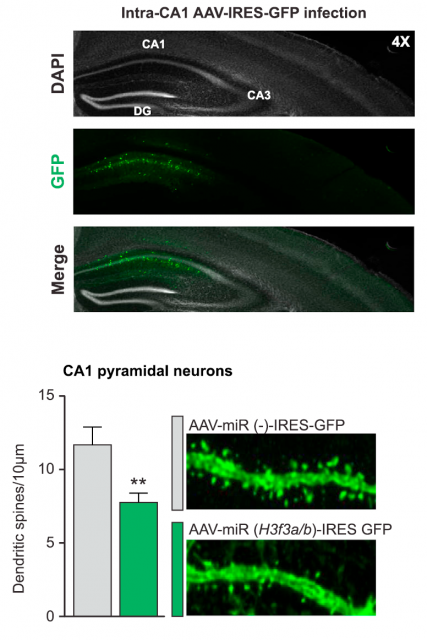
But a question remained unanswered: how is this fine mechanism of histone turnover regulated? The process is probably complex and involves more participants, but two of them were discovered: the chaperone Hira, implicated in chromatin remodeling, was necessary for a correct H3.3 recycling. And on the other hand, the activity of the proteasome, the macrostructure responsible for specific and controlled protein degradation inside cells, was also concomitant for a correct elimination of histone H3.3 after a forced neuronal activity. In summary: neuronal stimulation produces an increase in H3.3 expression, followed by a subsequent regeneration of the H3.3 bound to chromatin, which is substituted by the newly synthesized one. This dynamic recycling of H3.3 is a novel mechanism and is related to the neuronal stimulation; to get a final, general picture of the relevance for neuronal plasticity, there was something left: uncovering the effects of this recycling process unto the overall expression pattern of the neuronal genome. This was achieved using an obvious but effective strategy. Using microRNA-mediated silencing, the genes encoding H3.3 were knocked down, and the effect on neuronal life was precisely registered. Surprisingly, neurons could still be alive and apparently healthy; but they showed critical differences. For instance, the silencing of H3.3 affected dendritic spine number, which was further restored when adding back H3.3. A thorough bioinformatics analysis of the genes enriched with the most dynamic H3.3 exchange in adult mice showed a prevalence of genes previously related to cognitive processes and memory. To corroborate the physiological implications of these observations, exogenous impairment of the histone recycling in the adult mice demonstrated that exploratory behaviors and cognitive skills of the animals were significantly affected. The link between the molecular process and the functional expression of phenotype was finally confirmed.
The general understanding of brain plasticity has been, during the latest years, that create new and more numerous connections which are, in turn, responsible for the improvement of brain functions. Nowadays we have learned that neurons can proliferate, to some extent, and that the brain has some regenerative capacity in particular circumstances. But the mechanisms of learning and memory are poorly understood, and this is one of the reasons underlying our terrible delay in diagnosing, treating and finally healing mental illness that accounts with memory loss or cognitive impairment at different levels. The mechanism described in this work provides insightful clues and opens a very promising path: simply by recycling the pool of histones, new chromatin rearrangement can influence the way neurons connect to each other, without the need for a complex codification of histone postranslational modifications. If this mechanism is also present at cell-specification processes in other tissues, we might get to better understand the mechanisms of cell differentiation with the obvious benefits for modern cell engineering; but in any case, the most valuable use for this information concerns the part of ourselves that still holds the most intriguing secrets: the outstanding potential and plasticity of human mind.
References
- Maze I, Wenderski W, Noh KM, Bagot RC, Tzavaras N, Purushothaman I, Elsässer SJ, Guo Y, Ionete C, Hurd YL, Tamminga CA, Halene T, Farrelly L, Soshnev AA, Wen D, Rafii S, Birtwistle MR, Akbarian S, Buchholz BA, Blitzer RD, Nestler EJ, Yuan ZF, Garcia BA, Shen L, Molina H, Allis CD. Critical Role of Histone Turnover in Neuronal Transcription and Plasticity. Neuron. 2015 Jul 1;87(1):77-94. DOI: 10.1016/j.neuron.2015.06.014. ↩