How to study the protein corona using fluorinated nanoparticles
Author: Mónica Carril is an Ikerbasque Research Associate at the Biophysics Institute CSIC-UPV/EHU.
When nanoparticles (NPs) get in contact with biological fluids such as blood, proteins present in it will adsorb on the surface of those NPs forming what is known as the protein corona. It is a dynamic process in equilibrium with the surrounding proteins and it may lead to drastic changes in the NPs. The protein corona masks the surface of NPs and alters their physicochemical properties for which it is a matter of concern in the field of nanomedicine. If a NP is designed for a biomedical application with a particular charge, ligand or targeting moiety, the presence of the corona may alter size and surface properties, modifying unwantedly the fate and excretion pathways of the NPs in vivo, most likely shifting them away from the desired target. 1
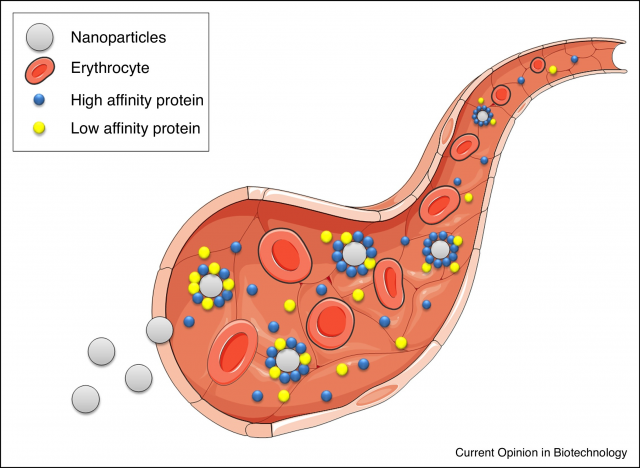
For these reasons, the protein corona has been extensively studied by multiple techniques. However, and in order to be analysed, NPs with protein corona usually have to be isolated from the protein solution leading to a loss of the equilibrium situation. Frequently, the protein corona is studied by measuring the size increase of the NPs in solution due to the layer of proteins attached onto them and one way to do so is by measuring the diffusion of NPs which can be correlated with their size. Several optical methods have been used to measure diffusion of NPs, however optical methods in complex media suffer from light scattering and cannot be used in turbid environments. [1] Of course, to be able to study protein corona formation in vivo and in real time, rather than trying to emulate it in the lab, would be major breakthrough. The use of non-optical methods such as Magnetic Resonance Spectroscopy (MRS) brings us closer to the actual in vivo and in situ protein corona formation evaluation.
In our paper,2 we describe diffusion measurements by Fluorine-based Nuclear Magnetic Resonance (19F NMR) spectroscopy as a non-optical based method which obtains diffusion information from fluorinated species without interference from the background, due to the natural absence of fluorine in biological fluids. We designed and prepared different water dispersible fluorinated NPs suitable for providing an adequate signal in 19F NMR. By exposing those fluorine-labelled NPs to mixtures of proteins, plasma, blood or cells, it was possible to measure their diffusion in equilibrium with the surrounding medium, without the need to isolate them.
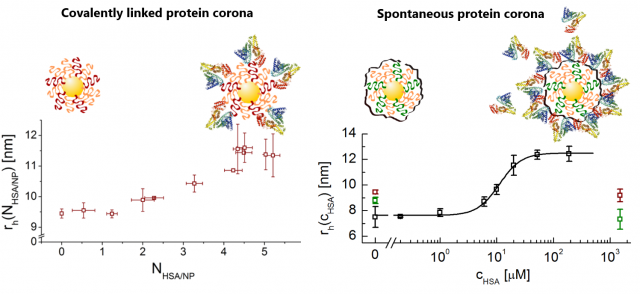
In a typical experiment, the signal intensity decay due to the diffusion of NPs is recorded by 19F-based diffusion-ordered nuclear magnetic resonance spectroscopy (DOSY) experiment, subsequently fitted to a mono-exponential decay to obtain a value for the diffusion constant (D), which is used to calculate the hydrodynamic size (rh) of the NPs via the Einstein-Stokes relation. Initially and as a proof of concept, artificial coronas were prepared by chemically attaching an increasing number of proteins onto the surface of selected fluorinated NPs. As the number of proteins increased, so did the size of the resulting NP-complex, as obtained from diffusion measurements by 19F NMR.
Next, we tested our methodology to evaluate the formation of non-covalent coronas, i.e. spontaneously formed coronas by exposing one type of our fluorine-labelled NPs to increasing amounts of single plasma proteins, such as human serum albumin (HSA) or transferrin (TF). Proteins adsorbed onto the surface of our NPs leading to a size (rh) increase detectable by our method.
Finally, measurements in more realistic complex media were performed. Thus, several 19F-labelled NPs were mixed with real samples of human blood and human plasma, and diffusion measurements were done at 37 ºC, to mimic as much as possible physiological conditions. We noticed that depending on the different surfaces of the tested NPs we had a very different response in each medium. We observed a consistent size shrinkage for some NPs, but in other cases a size increase was detected. Interestingly, in all cases the data obtained after incubation with HSA, the most abundant protein in human plasma, plasma or blood were different among them for the same NP type. These data suggest that in vitro protein corona studies are only simulations that may give us a hint on the tendency of our NPs to adsorb proteins on their surface or not, but are unable to model the in vivo protein corona.
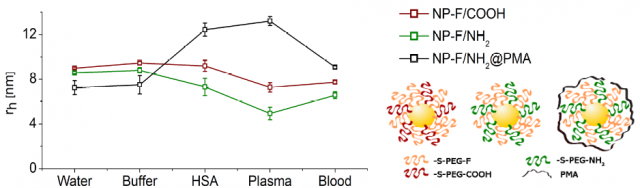
Hence, it is important to advance in the knowledge of the real protein corona as it influences the fate of nanomaterials in vivo and their applications as potential therapeutic and diagnosis nanotools. The use of magnetic resonance as a diffusion measuring technique opens up the possibility to measure NPs size in vivo using an MRI scanner. It must be noted that this method is exclusively based on measuring diffusion constants to obtain size, and is insensitive to information on a molecular level or the proteins involved in the corona. Obviously, we envisage that interpretation of changes in hydrodynamic radii in vivo will not be straightforward given the complexity of a living being. Nonetheless, these measurements are a promising starting point for future monitoring of the geometry changes of NPs in vivo.
References
- Carolina Carrillo-Carrion, Mónica Carril, Wolfgang J. Parak. Techniques for the experimental investigation of the protein corona. Curr. Opin. Biotech. 2017, 46, 106. doi: 10.1016/j.copbio.2017.02.009. ↩
- Mónica Carril, Daniel Padro, Pablo del Pino, Carolina Carrillo-Carrion, Marta Gallego, In situ detection of the protein corona in complex environments. Nat. Commun. 2017, 8, 1542. doi: 10.1038/s41467-017-01826-4. ↩