Design of protein-protein binding sites suggests a rationale for naturally occurring contact areas
Author: Ivan Coluzza is an Ikerbasque Research Professor at CICbiomaGUNE
Molecular recognition is a critical process for many biological functions and consists in non-covalent binding of different molecules, such as protein-protein, antigen-antibody and many others. The host-guest molecules involved often show a shape complementarity, and one of the leading specification for molecular recognition is that the interaction must be selective, i.e. the host should strongly bind to one selected guest and poorly, if at all, to all other biomolecules.
Here 1 we present a computational approach that simplifies the protein-protein system, reducing it to an artificial protein-binding site (BS), so to isolate the contribution due to the chemical heterogeneity and the steric compatibility on the selectivity power of the BS.
We developed a computational protein model that correctly represents the protein configurational space and the relation between sequence and structure. Such condition it is essential in order to study the the influence of the binding process on the evolution of the protein sequences. The latter in fact are not random and the order of the amino acids along the chain encodes for the folding and binding properties of the artificial protein.
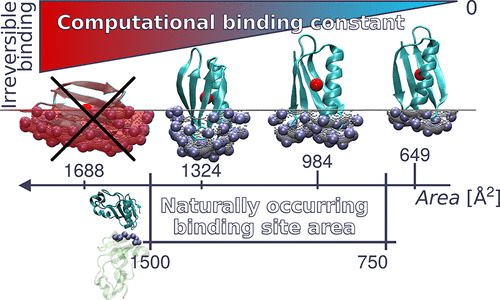
To this end we investigate four systems differing in terms of BS surface size. We design the target proteins to bind to their designated BS. The first observation is that all designed proteins sequences fold into the target structure both in bulk and when bound to the BS. Furthermore, the proteins correctly dock into the target position to their respective binding-site. Moreover, we demonstrate the specificity of the BSs of all sizes. It is important to stress that the specificity came about spontaneously from the design process and did not require any refinement step.
Hence, the first result of the present paper is that it seems that specificity is a direct consequence of the surface steric selection and the design algorithm. Such a result alone implies that the evolution of protein-protein interaction in the heterogeneous medium of living cells is a simpler optimisation problem, rather than of combinatorial complexity where thousands of random binding events are designed out.
The second crucial result that we present is the observation of the presence of an upper and a lower limits for the maximum and minimum area size available for a BS. Our data match the experimental observation of such limits
(750 -1500 Å2) and provide a rationale for them: the value of the binding constant limits the extent of the BS area. BS smaller than the identified range cannot offer binding affinities strong enough to withstand thermal fluctuations.
Larger BS, on the other hand, are not necessary as the in presented here already can be optimised to have enormous binding affinities. Such observations have far-reaching implications in the understanding of the evolution of protein-protein interactions and could be used to guide the design of artificial novel BSs with desired binding affinity and specificity.
References
- Francesca Nerattini, Luca Tubiana, Chiara Cardelli, Valentino Bianco, Christoph Dellago, and Ivan Coluzza (2018) Design of Protein–Protein Binding Sites Suggests a Rationale for Naturally Occurring Contact Areas Journal of Chemical Theory and Computation doi: 10.1021/acs.jctc.8b00667 ↩