Mechanodrugs
Mechanodrugs
Numerous proteins in the cell withstand mechanical loads while performing their function. This is especially significant for cell-surface proteins located in the extracellular matrix, which are essential for the communication between cells in the extracellular milieu. Reacting to mechanical force through conformational changes is crucial for these cell-surface proteins, translating a physical signal into an intracellular signalling process, or establishing physical connection with other cells.
Over 1400 cell-surface proteins compose the human surfaceome, including integrins, intercellular adhesion molecules, and cluster of differentiation molecules, which highlights the importance of protein mechanics in the cell. Similarly, viruses and bacteria use their own surface proteins to establish anchoring with cell-surface molecules to initiate infection. Again, the mechanical stability of these protein–protein interactions plays a crucial role in the success of the infection process, implying an important function of mechanical force in viral entry and bacterial adhesion. In fact, it is known that perturbing such interaction may result in avoidance of infection.
Protocols with different approaches have been designed to control the mechanostability of proteins; however, introducing mutations, using antibodies or metal ions, just to name some of them, may have some complications for practical implementation as mechanomodulators.
Now, a team of researchers proposes 1 a mutation-free approach to alter protein mechanostability using small molecules. The technology combines searching thousands of compounds from virtual chemical libraries similar to procedures commonly used in drug discovery, molecular docking, i.e., targeting specific regions previously known as potential relevant mechanical sites, and single-molecule atomic force spectroscopy (smAFS).
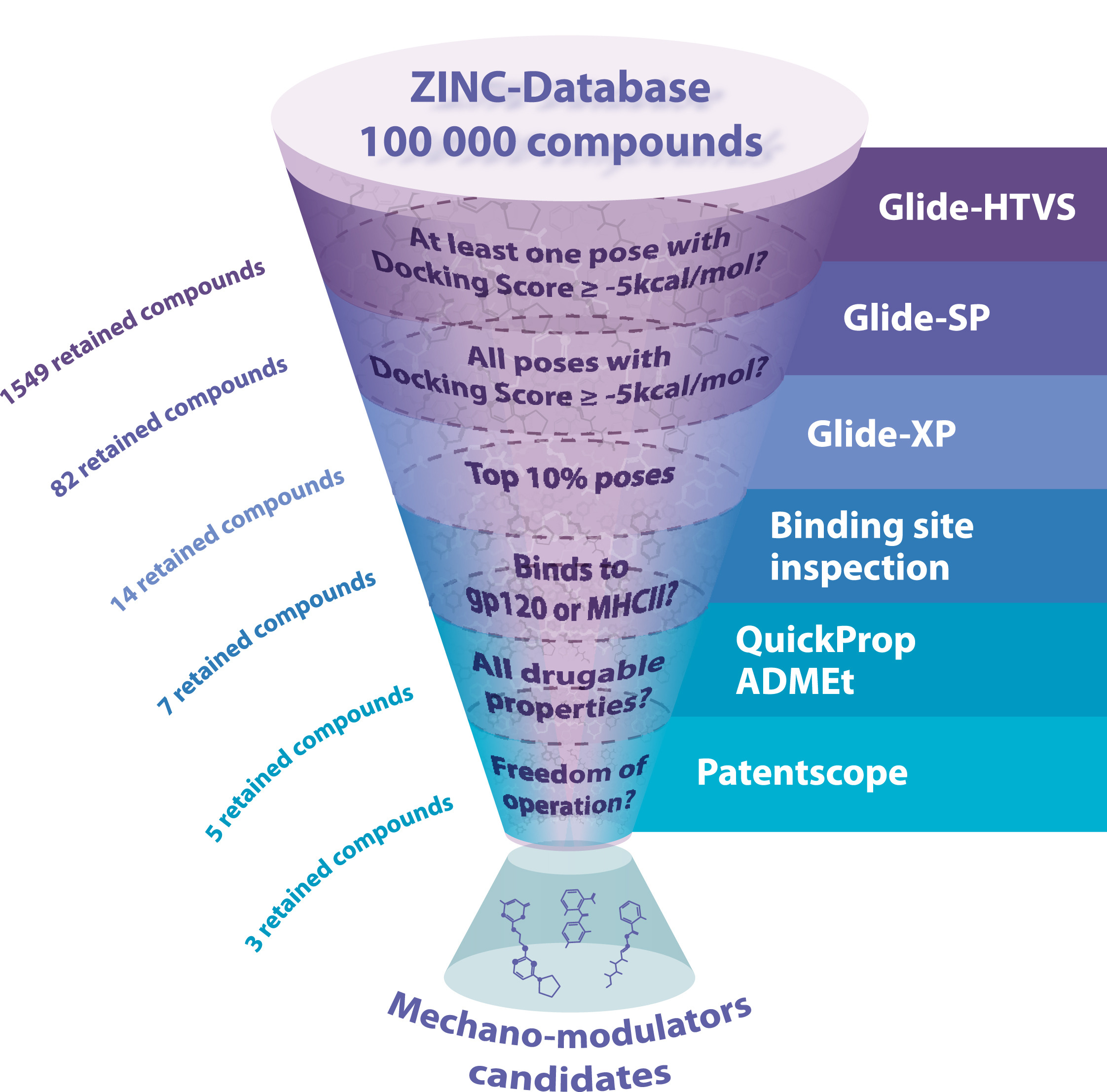
The team applies this approach to CD4 protein, a coreceptor present in T lymphocytes membrane, which is involved in antigen recognition, but also, it is the primary receptor of the human immunodeficiency virus HIV-1.
The researchers identified three small molecules in smAFS experiments, which probe their ability to modify and enhance CD4 mechanical stability, thus acting as protein mechanical stability regulators (PROMESRs). They propose that PROMESR might be useful molecules not only to alter the mechanical stability of cell-surface protein but also that of any protein whose function relies on its mechanical integrity. Thus, PROMESR may be useful to interfere with any protein–protein interaction process that occurs with the intervention of forces, such as those happening between microbes and host cells or cell–cell interaction.
These PROMESR molecules are a proof of concept of mechanoactive molecules discovered by means of a drug discovery pharmacology-based approach, bringing the possibility of a new type of molecular function, that is, mechanoregulation, and a new class of mechanodrugs.
Author: César Tomé López is a science writer and the editor of Mapping Ignorance
Disclaimer: Parts of this article may have been copied verbatim or almost verbatim from the referenced research paper/s.
References
- Antonio Reifs, Alba Fernandez-Calvo, Borja Alonso-Lerma, Jörg Schönfelder, David Franco, Mariano Ortega-Muñoz, Salvador Casares, Concepcion Jimenez-Lopez, Laura Saa, Aitziber L. Cortajarena, David De Sancho, Eider San Sebastian, Raul Perez-Jimenez (2024) High-throughput virtual search of small molecules for controlling the mechanical stability of human CD4 Journal of Biological Chemistry doi: 10.1016/j.jbc.2024.107133 ↩