DEK::NUP214 acts as an XPO1-dependent transcriptional activator of essential leukemia genes
DEK::NUP214 acts as an XPO1-dependent transcriptional activator of essential leukemia genes
Author: Marta Irigoyen is a postdoctoral researcher at CIC bioGUNE
Acute myeloid leukemia (AML) with t(6;9)/DEK::NUP214 is recognized as a separate entity in the World Health Organization classification of myeloid neoplasms, accounting for 1% of all AML cases 1 and characterized by a high relapse rate and young age at diagnosis 2. The t(6;9) chromosomal rearrangement results in the fusion of almost the entire peptide sequence of DEK with the C-terminal region of NUP214, encoding the chimeric protein DEK::NUP214 (2). NUP214 is a FG nucleoporin anchored to the cytoplasmic ring of the Nuclear Pore Complex that interacts with multiple transport receptors, including exportin 1 (XPO1).
These interactions are known to be maintained in DEK::NUP214 3. DEK is an ubiquitously expressed nuclear factor with multiple functions, including gene regulation 4. A recent study has shown that t(6;9) AML displays a gene expression signature reminiscent of other AML subtypes 5. However, the molecular mechanisms underpinning the pathogenicity of DEK::NUP214, and how these transcriptional changes are addressed remain unclear. In the present paper, through multi-omics and functional approaches in primary AML patient samples and human cellular models, authors observed a strong and selective sensitivity to XPO1 inhibition of t(6;9) cells and demonstrated that DEK::NUP214 acts as an XPO1-dependent transcriptional activator of essential leukemia drivers 6.
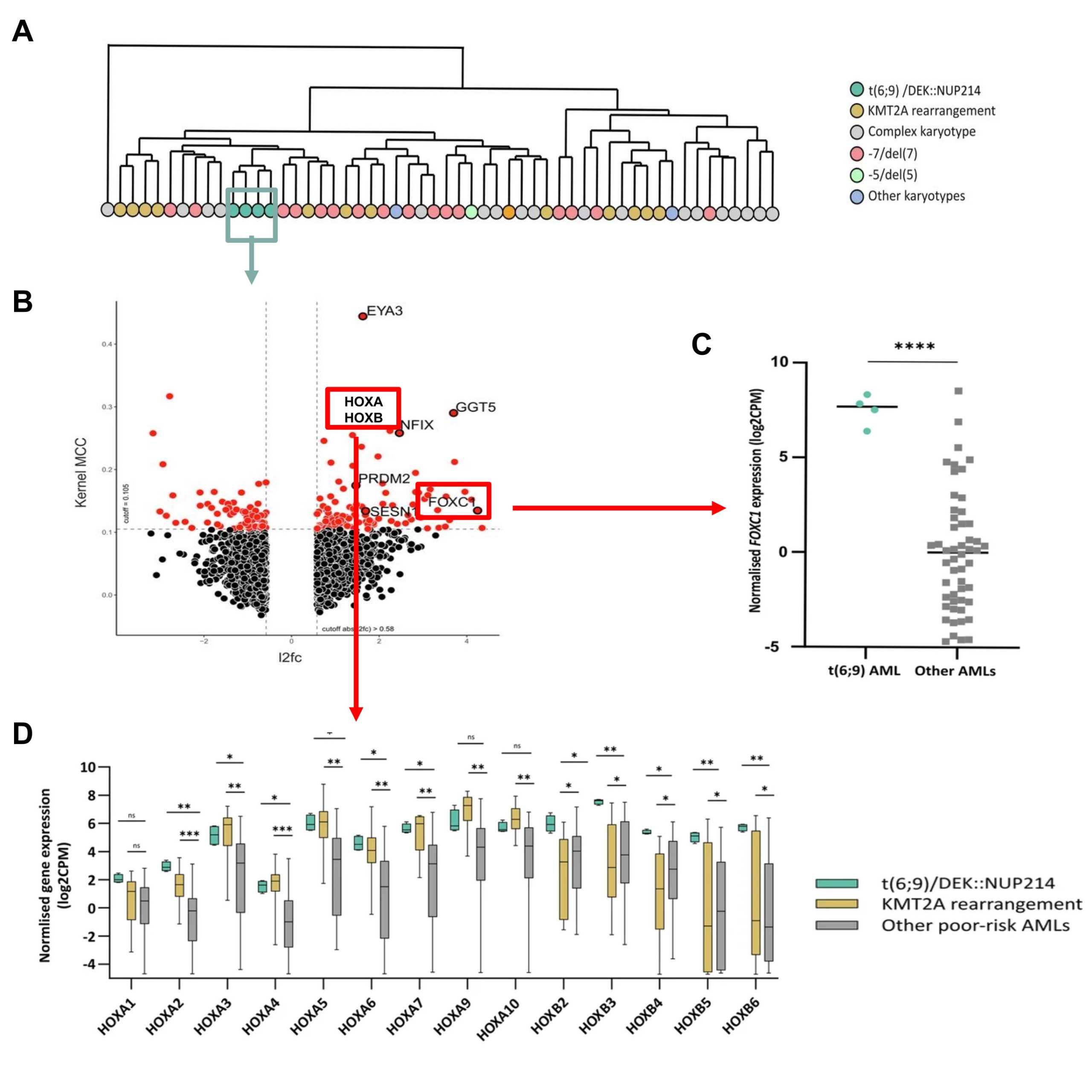
Source: Modified from Kaya, F. et al. (2025) Leukemia (2025). doi: 10.1038/s41375-025-02593-8. CC BY 4.0
To identify potential mediators underlying DEK::NUP214 leukemogenesis, authors firstly analyzed RNA-seq (a method for sequencing an entire set of RNA molecules) data from a cohort of 57 cytogenetically poor-risk AML samples that included four t(6;9) patients 7. The median age of these DEK::NUP214 cases was 43.5 years with a mean overall survival of 1.18 years. Thus, they first observed that AML samples from t(6;9) patients clustered distinctly from other AMLs, suggesting a unique transcriptional signature. To further explore this findings, they integrated their RNA-seq data with a separate series of 691 AML cases from all cytogenetic groups. Their analysis identified 166 differentially expressed genes in t(6;9) patients compared to other AML samples. Among the 104 significantly upregulated transcripts, they found known leukemia drivers (FOXC1 and HOXB genes), genes previously reported as being dysregulated in t(6;9) patients (5). On the one hand, HOXA and HOXB genes are a family of hematopoietic transcription factors whose deregulation is a feature of leukemogenesis. t(6;9) AML showed the overexpression of HOXA genes characteristic of other AML subtypes (5), whereas overexpression of HOXB genes appeared characteristic of t(6;9) samples. On the other hand, FOXC1 (another transcription factor) associated with poorer AML outcomes 8, was found strikingly overexpressed in t(6;9) cases. Given the established role of FOXC1 in AML pathogenesis, authors decided to analyze in more depth its involvement in FKH-1 cell lines (these cells are an AML cell line that harbors an identical t(6;9) translocation). Using shRNA-mediated silencing, they observed that the abolition of this gene led to a significant increase in apoptosis, cell cycle arrest, and a decrease in the number of colonies under hypoxic conditions (3% O2), paralleled to a significant downregulation of HOX genes expression.
Next, to identify therapeutic vulnerabilities of t(6;9) patients, authors carried out an in vitro drug screening in four primary samples using an established drug sensitivity and resistance-testing platform (DSRT) (7). Thus, they integrated these existing data with their results in t(6;9) samples to identify the most selective and effective compounds for these patients. This analysis revealed that two XPO1 inhibitors “Eltanexor” and “Selinexor, included in the drug screening, ranked in the top-4 compounds tested. Likewise, FKH-1 showed enhanced sensitivity to XPO1 inhibitors compared to AML cell lines with distinct cytogenetic backgrounds. In support of these findings, previous studies reported the interaction between DEK::NUP214 and XPO1 through the NUP214-FG domain 9. In order to investigate whether XPO1 inhibition was able to alter t(6;9)/DEK::NUP214 transcriptomic profile, they performed RNA-seq post in vitro exposure to Selinexor of t(6;9) and non-t(6;9) primary AML cells. Interestingly, 36 out of the 104 genes upregulated at diagnosis in t(6;9) primary samples as compared to other AML samples, including FOXC1 and HOX genes, were significantly downregulated in t(6;9) cells upon XPO1 inhibition, but, critically, not in non-t(6;9) primary samples. As expected, no differences were observed in the expression of these genes upon the control treatment in any of the samples analyzed.
Taking all this into account, their findings raised the question of whether DEK::NUP214 had a direct role in transcriptional regulation and whether XPO1 contributed to this effect. To address this, they first overexpressed HA tagged-DEK::NUP214 (HA-tag is a short amino acid tag added to the protein of interest) by lentiviral transduction of HEK-293T, an easy-to-transfect cell line, generating the DN-HA-293T model used to validate the interaction between DEK::NUP214 and XPO1 by co-immunoprecipitation. Next, they conducted CUT&RUN experiments (a method used to analyze protein interactions with DNA) in both the DN-HA-293T model (using an HA antibody) and the FKH-1 cell line (using a NUP214 antibody). These experiments revealed that DEK::NUP214 was able to bind to the promoters of 801 genes, with around one third encoding DNA or RNA binding proteins involved in transcription, DNA replication, DNA repair and RNA processing. Of note, 10 of the genes differentially expressed between t(6;9) and non-t(6;9) patients were also among the direct targets of DEK::NUP214 identified in the CUT&RUN experiments, including FOXC1 and HOXB genes. Besides, CUT&RUN experiments performed after Selinexor treatment of FKH-1 cells showed that XPO1 inhibition resulted in the loss of the binding of DEK::NUP214 to the regulatory regions of these target genes, indicating that this interaction was dependent on the presence of XPO1 and explaining the specific gene expression rewiring after Selinexor treatment in t(6;9) cells.
Altogether, authors demonstrate that a distinct transcriptional program is conserved in t(6;9) AML and characterized by the overexpression of essential leukemic factors (e.g., FOXC1, HOXA and HOXB genes) that are directly regulated by DEK::NUP214. They also found a functional interplay between XPO1 and the fusion protein, resulting in a selective sensitivity of t(6;9) cells to XPO1 inhibition that specifically reverts this transcriptomic profile.
References
- Vardiman JW, Thiele J, Arber DA, Brunning RD, Borowitz MJ, Porwit A, Harris NL, Le Beau MM et al. (2009) The 2008 revision of the World Health Organization (WHO) classification of myeloid neoplasms and acute leukemia: rationale and important changes. Blood doi: 10.1182/blood-2009-03-209262. ↩
- Slovak ML, Gundacker H, Bloomfield CD, Dewald G, Appelbaum FR, Larson RA, Tallman MS, Bennett JM et al. (2006) A retrospective study of 69 patients with t(6;9)(p23;q34) AML emphasizes the need for a prospective, multicenter initiative for rare ‘poor prognosis’ myeloid malignancies. Leukemia doi: 10.1038/sj.leu.2404233. ↩
- Saito S, Cigdem S, Okuwaki M, Nagata K. Leukemia-associated Nup214 fusion proteins disturb the XPO1-mediated nuclear-cytoplasmic transport pathway and thereby the NF-κB signaling pathway. Mol Cell Biol. 2016;36:1820-35. PMID: 27114368 DOIS: 10.1128/MCB.00158-16. ↩
- Riveiro-Falkenbach E, Soengas MS. Control of tumorigenesis and chemoresistance by the DEK oncogene. Clin Cancer Res. 2010;16:2932-8. PMID: 20501624 DOI: 10.1158/1078-0432.CCR-09-2330. ↩
- Potluri S, Kellaway SG, Coleman DJL, Keane P, Imperato MR, Assi SA, Cockerill PN, Bonifer C. Gene regulation in t(6;9) DEK::NUP214 acute myeloid leukemia resembles that of FLT3-ITD/NPM1 Acute Myeloid Leukemia but with an altered HOX/MEIS axis. Leukemia. 2024;38:403-7. PMID: 38172329 DOI: 10.1038/s41375-023-02118-1. ↩
- Kaya F, Bewicke-Copley F, Miettinen JJ, Casado P, Leddy E, Deniz Ö, Lavallée VP, Philippe C et al. Leukemia. DEK::NUP214 acts as an XPO1-dependent transcriptional activator of essential leukemia genes. Leukemia. 2025 Apr 9. PMID: 40204893 DOI: 10.1038/s41375-025-02593-8. Online ahead of print. ↩
- Casado P, Rio-Machin A, Miettinen JJ, Bewicke-Copley F, Rouault-Pierre K, Krizsan S, Parsons A, Rajeeve V et al. Integrative phosphoproteomics defines two biologically distinct groups of KMT2A rearranged acute myeloid leukaemia with different drug response phenotypes. Signal Transduct Target Ther. 2023;8:80. PMID: 36843114 DOI: 10.1038/s41392-022-01288-1. ↩
- Simeoni F, Romero-Camarero I, Camera F, Amaral FMR, Sinclair OJ, Papachristou EK, Spencer GJ, Lie-A-Ling M et al. Enhancer recruitment of transcription repressors RUNX1 and TLE3 by mis-expressed FOXC1 blocks differentiation in acute myeloid leukemia. Cell Rep. 2021;36: 109725. PMID: 34551306 DOI: 10.1016/j.celrep.2021.109725. ↩
- Sandahl JD, Coenen EA, Forestier E, Harbott J, Johansson B, Kerndrup G, Adachi S, Auvrignon A et al. t(6;9)(p22;q34)/DEK-NUP214-rearranged pediatric myeloid leukemia: an international study of 62 patients. Haematologica. 2014;99:865-72. PMID: 2444114 DOI: 10.3324/haematol.2013.098517. ↩