LUCA, a prokaryote-like organism at war with viruses
LUCA, a prokaryote-like organism at war with viruses
Author: Saioa Manzano-Morales, PhD candidate at the Comparative Genomics group at the Barcelona Supercomputing Center (BSC)
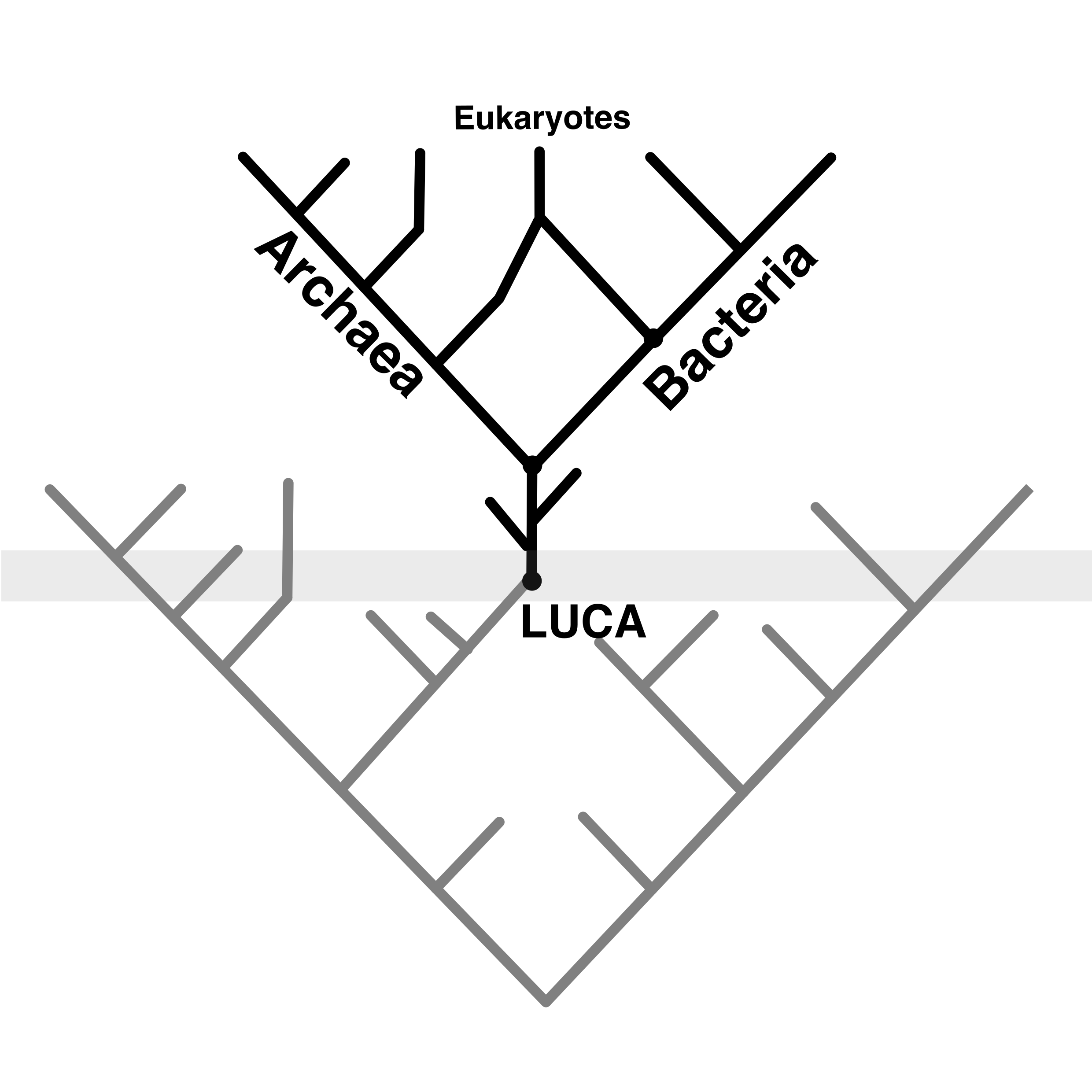
All cellular life as we know it today stems from a single common ancestor, from which all modern lineages emerged. The origin of cellular life is thought to be the node in the Tree of Life from which Archaea and Bacteria, the two primary domains, diverged.
The universality of some features across cellular life evidences the existence of a Last Universal Common Ancestor (LUCA), but there is considerable discrepancy on what LUCA looked like, the kind of environment in which it lived, and when. Understanding the earliest ancestor that binds all cellular life together is key to understanding the early evolution of life on Earth, and therefore of all that came after.
A recent study 1 sheds light on the age and putative metabolism and ecology of LUCA, by combining state-of-the-art evolutionary inference methods and biogeochemical models of the early Earth.
A molecular clock for LUCA
Inferences on past species are usually made through the fossil record. However, it is sparse, particularly so for unicellular organisms, which do not have hard structures that are readily fossilizable. On the contrary, protein sequences–encoded in the DNA–are an abundant source of evolutionary information, as they are bookkeepers of all the changes (that is, mutations) that have occurred to them during evolution. By comparing sequences across a wide set of organisms, and employing models that capture these evolutionary changes through time, evolutionary biologists can infer the processes that have driven their evolution, and reconstruct the relationships of the organisms that encode them in their genomes. Considering also that these sequences mutate as a product of time, they can additionally be leveraged to translate this sequence divergence into units of time (a theory called Molecular Clock).
Some gene families are more agreeable to molecular clock analyses than others. By employing protein families that have essential functions in the cell, and that duplicated before LUCA (namely, the catalytic and non-catalytic subunits of ATP synthase, elongation factor Tu and G, signal recognition protein and signal recognition particle receptor, tyrosyl-tRNA and tryptophanyl-tRNA synthetases, and leucyl- and valyl-tRNA synthetases), Moody et al. were able to reduce the inherent uncertainty in the inferences (as the same calibrations can be applied at least twice) and obtained an age estimate for LUCA ranging from 3.94 Ga to 4.52 Ga, in line with other studies.
LUCA was already a fairly complex organism
Beyond its age, by reconstructing the evolutionary relationship of 350 Archaea and 350 Bacteria, they then applied a probabilistic framework that estimates the evolution of all gene families in the Kyoto Encyclopedia of Genes and Genomes (KEGG), termed KEGG Orthologs (KOs). This allows mapping the presence/absence of these gene families in all ancestors of modern prokaryotes, up to and including LUCA, at a given probability.
These resulting probabilities allow us to account for uncertainty when reconstructing the genome of an organism that lived so far in the past. This reconstruction, though likely incomplete, provides a glimpse into this extinct organism, the same way that a partial dinosaur can provide useful information on its morphology, diet and behavior. Using modern prokaryotes as a training set, they estimated the relationship between KEGG KOs and genome size/total number of proteins, yielding a conservative estimate of around 2,600 proteins and a genome size of at least 2.5 Mb. This is comparable to modern prokaryotic cells, suggesting that LUCA was already a fairly complex organism.
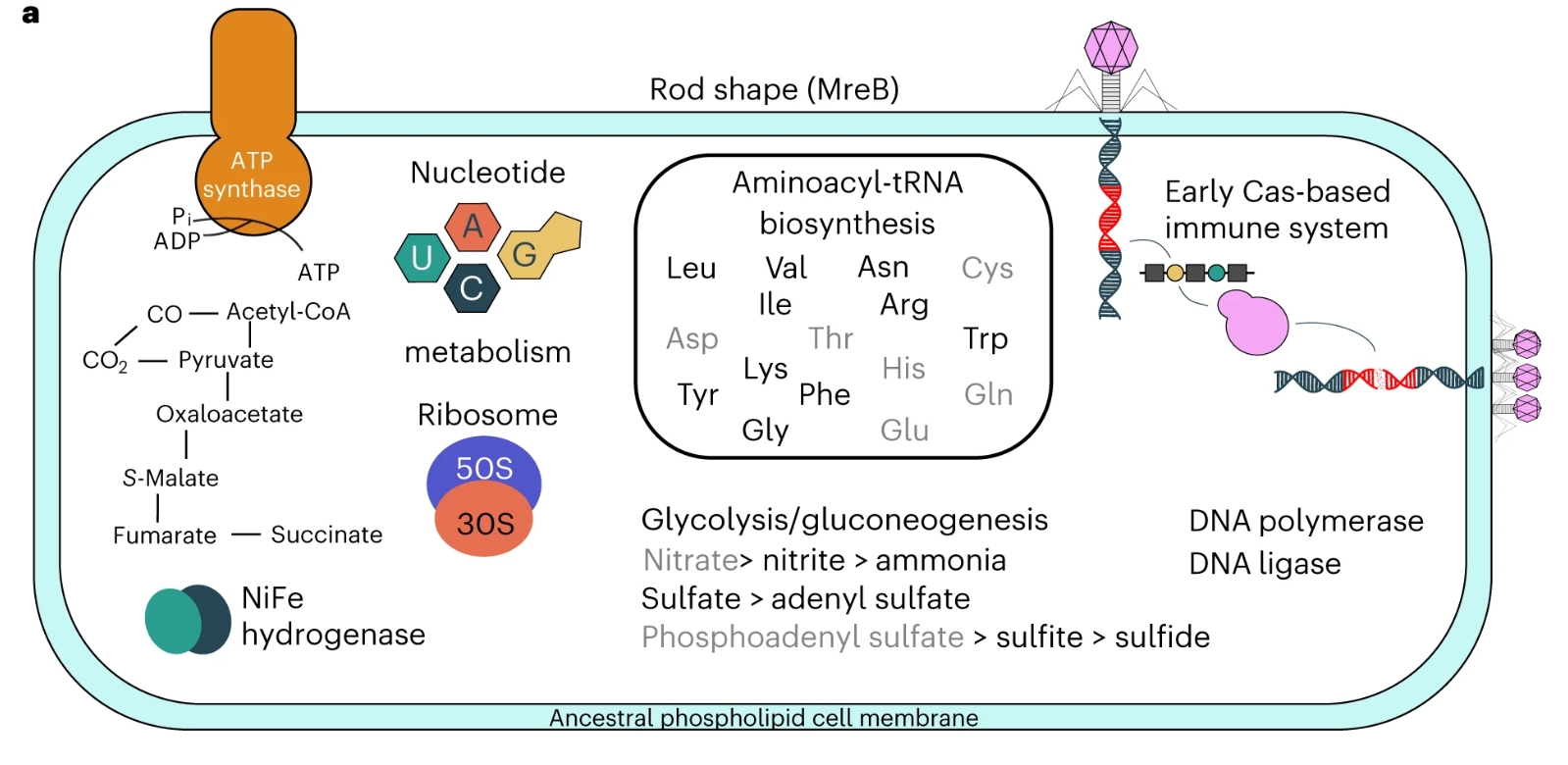
Analysis of the enzymatic pathways formed by these proteins can also inform hypotheses on the metabolism that LUCA could have lived on: the lack of enzymes for employing oxygen as an electron acceptor suggests that LUCA was an anaerobe, with the ability to fixate carbon and/or acetogenic growth as suggested by the presence of key pathways for this metabolism.
This, along with enzymes for gluconeogenesis and glycolysis, suggests that LUCA was probably capable of organoheterotrophic (employing organic compounds as electron donors, and obtaining organic carbon from the environment) and possibly chemoautotrophic (with the ability to generate organic carbon from abiotic sources, but not through the use of sunlight as energy source) growth, utilizing hydrogen as an electron donor.
Cellular life at war
Aside from its metabolism, researchers also found support for the presence of CRISPR-Cas protein families, which mediate adaptive immune response against viruses in extant prokaryotes. This means that cellular life was already at war with viruses at the time of LUCA, and therefore that viruses are at least as ancient as LUCA.
It is easy to envision LUCA living in isolation, but it is much more likely that it formed part of a complex ecosystem with other contemporary organisms that have not yielded modern lineages that survived to this day. Since, by definition, LUCA is the oldest ancestor that can be traced back starting from modern cellular life, we cannot access information on the nature and metabolism of these other contemporaries by comparing the genomes of species alive today.
LUCA’s ecosystem and environment
However, LUCA’s metabolism provides a glimpse into the kind of ecosystem it could have lived on, and therefore of the kind of ecological interactions such an ecosystem could have sustained. Acetogenesis has a low energy yield, which suggests an energy-limited early biosphere. Organoheterotrophic growth would require presence of autotrophic organisms to provide the organic compounds, and even a chemoautotrophic LUCA is unlikely to have lived in isolation as the by-products of its metabolism would have attracted other metabolisms (as is the case in modern ecosystems).
A reconstruction of LUCA’s metabolism can also hint at the kind of environment it could have lived in: either the deep ocean (where hydrothermal vents could have provided a source of hydrogen) or the ocean surface, where hydrogen would be provided from the atmosphere.
References
- Moody, E.R.R., Álvarez-Carretero, S., Mahendrarajah, T.A. et al. (2024) The nature of the last universal common ancestor and its impact on the early Earth system. Nat Ecol Evol 8, 1654–1666 doi: 10.1038/s41559-024-02461-1 ↩