Environmental DNA: Biodiversity data (like love) is in the air
Environmental DNA: Biodiversity data (like love) is in the air
Author: Saioa Manzano-Morales, PhD candidate at the Comparative Genomics group at the Barcelona Supercomputing Center (BSC)
DNA sequencing is getting cheaper than ever. This, coupled with advances in speed and portability, are allowing us to apply deep sequencing beyond the lab to environmental substrates, and analyse this eDNA to gain information and monitor biodiversity at a time where it is being lost at an unprecedented rate. This environmental DNA can be obtained from substrate samples (e.g soil, water or sand), biological samples (e.g feces from the population of interest) or even air, which contains both microbes as well as DNA shed by multicellular organisms.

The analysis of airborne DNA has previously been restricted to targeted approaches: DNA metabarcoding, in particular, leverages the fact that some genes are highly conserved across species, while containing variable regions that allow species-specific identification. In this way, DNA probes are designed for the conserved regions to serve as primer for the DNA amplification, and subsequent sequencing allows for the identification based on the more variable regions. However, this has one key caveat, as it requires certain a priori sequence knowledge, and is therefore restricted to a given number of species of interest for which we can tailor the DNA probes, making invisible the species lacking the marker gene or for which the DNA probe is less suited.
Whole-genome approaches, which utilize random primers and sequence random portions of the genome until the fragments cover the whole genome, can be applied to multi-species samples (an approach called shotgun sequencing). These DNA sequences can then be taxonomically classified via a bioinformatics pipeline to assign them to species, with the relative counts of DNA per species being reflective of their relative abundance in the sample. This has the added benefit of being able to discern not only who is there but also what they’re doing, as the genes encoded in these DNA sequences can also be functionally annotated to discover, the prevalence of some biological traits of interest (for example, the presence and abundance of antimicrobial resistance genes within an environment to monitor the spread of resistance among bacteria).
This method has already been put in use for gut microbiome analysis, for example, and shotgun sequencing applied to environmental DNA has been tested on humans, microbes, and some multicellular species.
However, a recent paper 1 has gone a step further, showing that shotgun sequencing of airborne DNA is fast, reliable, scalable, and can be applied to a pan-domain-of-life scale, bringing us closer to real-time assessment of the community composition of an environment.
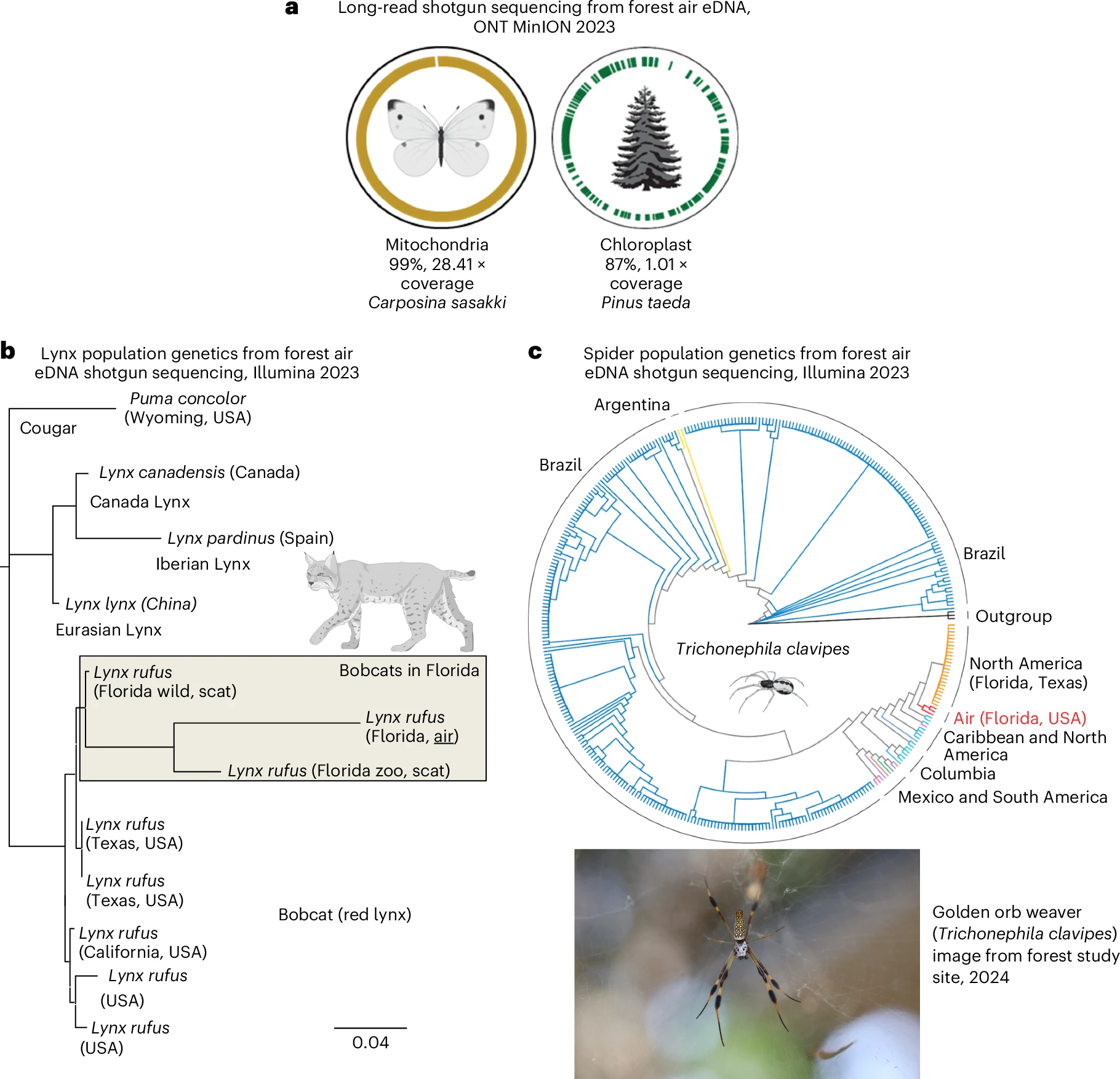
In the first pan-domain-of-life long-read analysis, they collected airborne DNA from several sampling sites (a total of 78 shotgun datasets) and applied it to a broad range of downstream applications:
- Non-invasive monitoring of biodiversity Tracking of species abundance through eDNA is particularly useful for species that are difficult to study through traditional methods, such as those that are cryptic, nocturnal, predators, or aerial. The study shows that, despite an overall positive correlation between the results of shotgun sequencing and metabarcoding, the shotgun approach reflects more accurately the original proportions of DNA in the sample and helps overcome some biases of metabarcoding such as barcode and taxonomic biases.
- Assessment of genetic diversity within a population of interest (powering population genetics and genetic variant analysis). This was tested both with humans, wild predators (bobcat) and arthropods (silk orb weaver spider).
- Pathogen and antimicrobial resistance surveillance, not limited to human pathogens but scalable to pathogens of agricultural or industrial importance.
- Disease vector and pest control. By detecting DNA of vectors of disease, such as black rats or mice, as well as arthropods like cockroaches and termites, this opens doors for large-scale pest surveillance.
- Environmental allergen scanning, such as peanuts or tree pollen.
Overall, these new eDNA technologies are a cost-effective and scalable approach for the non-invasive monitoring of biodiversity, as well as having applications for agriculture, biosecurity and medical applications. With near real-time detection of airborne DNA, we may be closer than ever to making Star Trek’s tricorder come true.
However, this emerging technology calls for regulations to establish the collection of human genetic information from environmental samples. The depth and accuracy to which human data can be gathered from airborne DNA raises ethical concerns about the possibility of individual-level surveillance that will need to be addressed, particularly if samples are taken from environments with high human habitation. Suggested targeted policies include regulations of who should directly investigate human eDNA and where such an investigation can take place, as well as what kind of approvals are needed, or molecular-based blocking of the human eDNA before processing and/or filtering of human-aligning reads out of the datasets before they are visible to investigators or submitted to public repositories.
References
- Nousias, O., McCauley, M., Stammnitz, M.R. et al. Shotgun sequencing of airborne eDNA achieves rapid assessment of whole biomes, population genetics and genomic variation. Nat Ecol Evol 9, 1043–1060 (2025). doi: 10.1038/s41559-025-02711-w ↩