Haplotypes, mitochondria and autism
Haplotypes, mitochondria and autism
An allele is each of the alternative forms that the same gene can have. The alleles differ in their DNA sequence and can manifest in different results after expressing that gene, such as differences in the color of the eyes or in the blood group. Human beings are diploid, we have two sets of chromosomes, one of them from the father and the other from the mother, and each pair of alleles is located in the same place or locus of each chromosome. A haplotype is a group of alleles from different places (loci) of a chromosome that are inherited together. A haplogroup is a large group of haplotypes that are inherited together and that share a common ancestor. They are a single hereditary line. Haplogroups are usually named with a capital letter and the most studied are those related to the Y chromosome (paternal line) and those related to mitochondrial DNA (maternal line). The haplogroups allow us to define the genealogical tree of humanity.
All the cells of animals have mitochondria in their interior, small organelles that are responsible for producing energy for the cell. The toxicity of cyanide is because it blocks the electron transport chain and prevents mitochondrial function. The production of cellular energy stops abruptly. When the ovule and the sperm are fused, the mitochondria are provided by the ovule and if there is some mitochondrial DNA from the father it is destroyed. However, in 2018 there has been a surprising result, at least 17 people have been found to have paternal mitochondrial DNA 1. We do not know how or why, but this discovery is forcing us to rewrite the books of human genetics. Researchers who have discovered this biparental mitochondrial inheritance believe that heteroplasmy, the diversity of mitochondria in the same cell, is due to the mutation of a gene in the nucleus, not in the mitochondria. The nuclear genome coordinates the process of destruction of the cellular components that are left over after fertilization is complete, including the paternal mitochondria that may have arrived with the sperm. The effects on the development of the embryo that heteroplasmy can have is unknown, we will have to rethink the effects of mitochondrial replacement therapies and we have to understand why in some cases the mtDNA of the father survives fertilization and how it does it.
A person’s DNA has therefore two main locations: the cell nucleus and the mitochondria. Nuclear DNA (nDNA) analysis of people with autism has found many variations in the number of copies (VNC) of heterozygous genes as well as mutations with loss of function, most of which explain only a few cases of ASD, which suggests a polygenic origin of ASD; that is, there is no “autism gene” but hundreds of genes that give more or less susceptibility to this disorder.
Until now we did not know anything about mitochondrial DNA (mtDNA) and TEA but that is changing. The metabolic profiles of people with ASD are consistent with mild mitochondrial defects and alterations have been seen in oxidative phosphorylation, the main mitochondrial function, basic for obtaining energy for the organism, in different cells and organs including lymphocytes, muscle and brain of people with ASD.
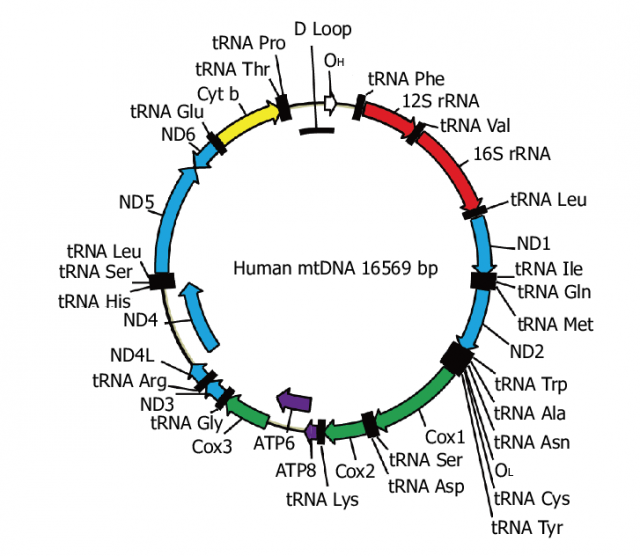
The mitochondrial genome is a double-stranded, circular, closed molecule, similar to bacterial DNA. Each cell has hundreds to thousands of copies of that mitochondrial DNA, each with 37 genes. These mtDNA genes encode 13 of the most important polypeptides of oxidative phosphorylation plus ribosomal RNA and transfer RNA for these genes to be transcribed in the mitochondria themselves. Therefore, mtDNA variants directly affect the cell’s energy metabolism.
One of the most important variations of mtDNA are the so-called ancient haplogroups. The history of humanity by matrilineal way can be reconstructed through these groups that are considered maternal lineages. The first modern humans, emerged in Africa, belong to the macrohaplogroup L. The most recent common ancestor has been called mitochondrial Eve, a woman who would have existed about 190,000 years ago and live in what is now Tanzania. In the following millennia, migrations and diversification would take place and different haplogroups can be distinguished. The current European population can be classified into seven classes, each of which would come from a single woman and which has been called “the seven daughters of Eve.” The first European «Eva» lived in present-day Greece about 45,000 years ago. The second lived in the Caucasus 25,000 years ago; the third about 20,000 years ago in Tuscany and the fourth about 17,000 years ago in Cantabria; the fifth lived in the area of the Pyrenees 17,000 years ago; the sixth appeared in central Italy 15,000 years ago and the seventh came about 8,500 years ago in Syria. Except for the Lapps, all the rest of the current European population results from those seven clans.
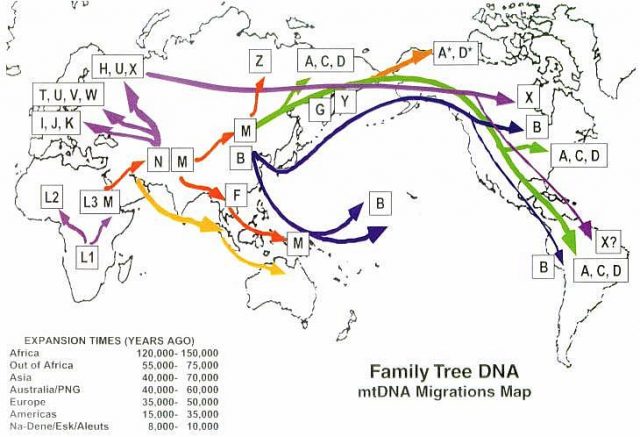
The differences in the haplogroups also allow to follow the geographical variations and the result of the migrations. About 65,000 years ago women carrying two variants of mtDNA (M and N) left Africa and their offspring settled in Eurasia, giving rise to the macrohaplogroups M and N. The macrohaplogroup M settled in Europe and from it derived haplogroups HHV, J, T, U, K (Uk), I, W and R. Other groups belonging to the macrohaplogroups M and N settled in Asia and generated 49% and 51% of the mtDNAs of Asians, respectively. The Asian macrohaplogroup M gave rise to different sublineages of M (M1 to M80) which include the haplogroups C, D, G and Z, while the Asian macrohaplogroup N gave rise to the Asian lineages A, B, F, O and others. The Eurasian haplogroups A, B, C, D and mtDNA C passed to the Americas. Since mtDNA does not recombine, all single nucleotide variations of a mtDNA lineage are imbalanced and function as a unit. Since the mtDNA of a mother, unlike its DNA, does not recombine with that of the father, it constitutes a certificate of genetic identity for each maternal line of the human genealogy.
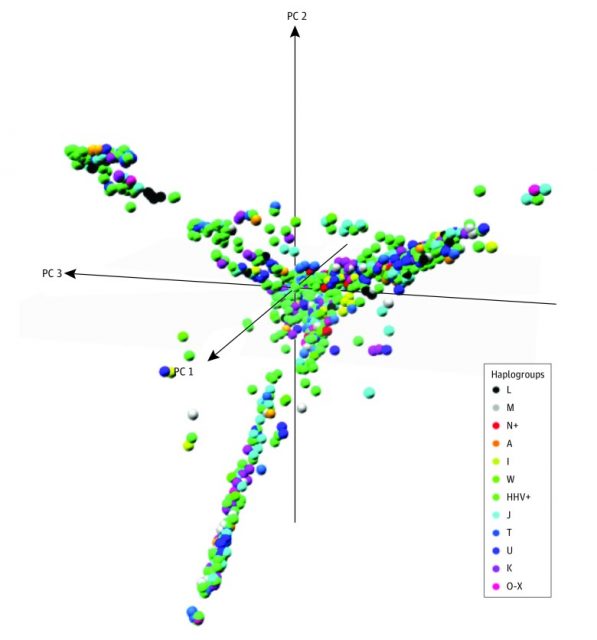
The different strains of mtDNA haplogroups are born and radiate from regional indigenous populations and are functionally different, which makes their proliferation within specific environments due to an adaptive selection. Therefore, defining a haplogroup also defines its functional variants. Although these variants are adapted to a specific regional environment, changes in the environment, such as migrations, changes in diet, or the influence of new infectious agents, can hinder that adaptation of a particular mtDNA lineage and result in phenotypes with clinical value. That is, some haplogroups may have a greater tendency to suffer from a disorder, particularly if it presents a link with mitochondrial alterations as occurs in autism.
Given the genetic complexity of ASD and the association between autism and alterations in mitochondrial bioenergetics, variation in mtDNAs, as expressed in haplotypes, may be a risk factor for autism. There are contrasted data that certain mutations in some of the 37 mitochondrial genes produce rare diseases.
The study by Chalkia and collaborators 2 used the Autism Genetic Resource Exchange database and took information from 933 families that included 4041 individuals, 1624 with ASD (80% boys and 20% girls) and 2417 parents and siblings without ASD. There were no restrictions on age, sex, racial profile or socioeconomic status.
The main result is that, compared to haplogroup HHV, the most common in Europe, the European haplogroups I, J, K, OX, T, and U were associated with an increased risk of suffering from an ASD, as was the case with Asian and native Americans A and M, with increases going, counting 1 as a reference value (HHV), from 1.55 to 2. Since those European haplogroups are expressed by 55% of the current European population, mtDNA lineages make a significant contribution to the risk of suffering an ASD. Therefore, neither the world population nor the entire European population has the same risk of autism.
References
- Luo S, Valencia CA, Zhang J, Lee NC, Slone J, Gui B, Wang X, Li Z, Dell S, Brown J, Chen SM, Chien YH, Hwu WL, Fan PC, Wong LJ, Atwal PS, Huang T (2018) Biparental Inheritance of Mitochondrial DNA in Humans. Proc Natl Acad Sci U S A 115(51): 13039-13044. doi:10.1073/pnas.1810946115 ↩
- Chalkia D, Singh LN, Leipzig J, Lvova M, Derbeneva O, Lakatos A, Hadley D, Hakonarson H, Wallace DC (2017) Association Between Mitochondrial DNA Haplogroup Variation and Autism Spectrum Disorders. JAMA Psychiatry 74(11): 1161-1168. doi: 10.1001/jamapsychiatry.2017.2604. ↩
2 comments
[…] Nuestro conocimiento de la relación entre el ADN mitocondrial y el autismo ha dejado de ser inexistente. Y este nuevo conocimiento enmarca esta relación en la historia genética de la humanidad. José Ramón Alonso en Haplotypes, mitochondria and autism […]
“Risk” of Autism? It’s not a disease! The comorbids can be, but Autism alone is just a neurodivergence.