Postzygotic mutations in autism
The number of genes linked to autism has not stopped growing in the last decade. Although there are hundreds of variants that are more frequent in people who are diagnosed with autism spectrum disorders (ASD) than in the general population, there are sixty genes that have a clearer linkage and could help build the genetic profile of autism or, most likely, the genetic profiles of the autisms. In fact, although we know that both genetic and environmental factors contribute to the aetiology of autism, genetic risk factors currently account for only a fraction of cases of ASD and much of the genetic aetiology of autism remains unknown.
The sequencing of genes in children with ASD and the comparison with the genomes of the father and mother (trios) have allowed identifying that many of these changes are de novo mutations; that is, they appear spontaneously, they are present neither in the father nor in the mother and they are not inherited.
In humans, DNA replication is estimated to entail one error in every 108 base pairs, giving rise to 30–100 genome-wide de novo mutations in each new generation. Whereas neutral or benign de novo point mutations contribute to normal genetic variation, single detrimental de novo mutations have been established to cause a number of rare developmental disorders and are increasingly recognized as a major contributor to common sporadic disorders, such as intellectual disability and autism. Several recent studies (1-3) have identified a new component of autism genetics: de novo mutations generated after conception, which are called somatic mutations or post-zygotic mutations. Cancer is predominantly caused by post-zygotic mutations. The peculiarity is that these mutations will appear only in the genome of part of the cells of the organism, forming what is known as a mosaic. Logically, if the mutation appears very early when the embryo is just a poorly differentiated cell group called a blastula, many cells in the mature organism will present the mutation. In contrast, as the mutation occurs in more advanced stages of embryonic development, fewer and fewer cells will carry that mutation and will, therefore, be more difficult to detect. How detrimental a post-zygotic mutation is to a person is dependent on what the mutation is, where it occurred in the genome and when it occurred.
Post-zygotic mutations can be important and silent elements. They can be small mutations that affect a single base pair or large mutations that affect entire chromosomes and are divided into two classes, spontaneous mutations and induced mutations. Induced mutations are caused by mutagens and they often demonstrate mutational specificity, meaning they cause predictable changes in the DNA sequence. A few common mutagens are ultraviolet light, base analogues, intercalating agents such as daunorubicin, reactive oxygen species and alkylating agents. F
Freed and Pevsner explored 1 the contribution of mosaic single-nucleotide variations to ASD using the whole exomes of 2,388 families. They found a strong bias from mosaic mutations in ASD probands relative to their unaffected siblings (1). Now, Dou et al. 2 have studied a similar number of families (2,361) and identified 1,248 putative post-zygotic single-nucleotide mosaicisms and 258 de novo single nucleotide polymorphisms in children with detectable parental mosaicisms (2). The estimation of these authors is that these post-zygotic mutations or de novo mutations inherited from parental post-zygotic mutations increased the risk of ASD by approximately 6%. In addition, this model revealed 13 new ASD risk genes.
The work done by Lim and his group focused 3 in the largest collection of trios (5,947) with ASD and performed resequencing using multiple independent technologies (3). Genomes were courtesy of Simons Foundation Autism Research Initiative (SFARI) Simplex Collection, the Autism Sequencing Consortium and Autism Speaks. This material, of public access, comes from autopsies of brain samples of people of different ages and deceased in different circumstances. Comparing this information on neuronal genetics with other sequencing data they found that 7.5% of the de novo mutations of people with ASD were post-genetic mutations. Of these, 83% had not been detected in the original genome analysis of these individuals.
Some of the genes affected by post-genetic mutations were already identified in that group of those linked to autism or other neurodevelopmental disorders, such as SCN2A, HNRNPU and SMARCA4, but some were affected in a different way to that known. In other cases, such as KLF16 and MSANTD2, these genes are known to be activated in brain development but had not been previously associated with autism. In early genetic studies, research focused on genes that involved a loss of function, usually the lack of the encoded protein or the production of a damaged, altered protein. However, the analysis of post-zygotic mutations shows that some of them imply an increase in expression, not a loss.
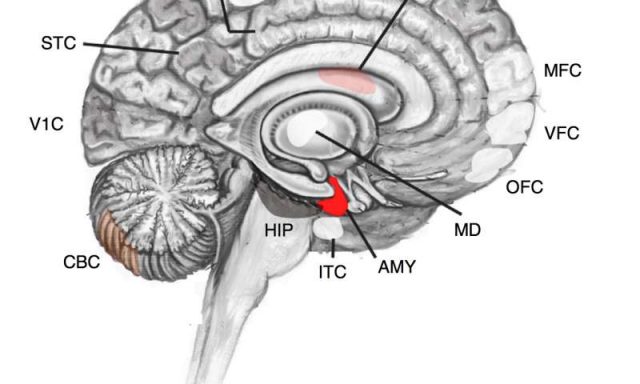
The use of another large database, the BrainSpan Project allowed for the analysis of post-zygotic mutations, the moment of origin of the mutations and affected brain regions. The research group found that the largest number of these mutations was found in the amygdala and to a lesser extent in the striatum and cerebellar cortex, where the difference between people with autism and controls did not reach statistical significance. It is a very interesting result because post-genetic mutations in people with ASD appear disproportionately high in genes expressed in the amygdala, which in turn is considered a key region in autism.
Post-zygotic, non-hereditary mutations appear to be important in various brain problems including epilepsy, intellectual disability, schizophrenia, and brain malformations. Autism is now added to that list.
References
- Freed D, Pevsner J (2016) The contribution of mosaic variants to autism spectrum disorder. PLoS Genetics, 12(9): e1006245. ↩
- Dou Y, Yang X, Li Z, Wang S, Zhang Z, Ye AY, Yan L, Yang C, Wu Q, Li J, Zhao B, Huang AY, Wei L (2017) Postzygotic single-nucleotide mosaicisms contribute to the etiology of autism spectrum disorder and autistic traits and the origin of mutations. Hum Mutat. 2017 Aug;38(8): 1002-1013. doi: 10.1002/humu.23255 ↩
- Lim ET, Uddin M, De Rubeis S, Chan Y, Kamumbu AS, Zhang X, D’Gama AM, Kim SN, Hill RS, Goldberg AP, Poultney C, Minshew NJ, Kushima I, Aleksic B, Ozaki N, Parellada M, Arango C, Penzol MJ, Carracedo A, Kolevzon A, Hultman CM, Weiss LA, Fromer M, Chiocchetti AG, Freitag CM; Autism Sequencing Consortium, Church GM, Scherer SW, Buxbaum JD, Walsh CA (2017) Rates, distribution and implications of postzygotic mosaic mutations in autism spectrum disorder. Nat Neurosci 20(9): 1217-1224. doi: 10.1038/nn.4598 ↩
2 comments
[…] Autismoaren oinarri genetikoak apurka-apurka eta zailtasun handiz argitzen ari dira. Agian zailtasun hau ematen ari da oinarri genetiko horiek ez daudelako ohikoa den tokian. Jose Ramon Alonsok azaltzen digu: Postzygotic mutations in autism. […]
[…] poco a poco y con gran dificultad. Quizás porque no están donde suelen estar. Postzygotic mutations in autism por JR […]