Do you want to make a symbiosis with me?
Do you want to make a symbiosis with me?
Symbiosis is a close relationship between two organisms that benefits both of them. Many different types of symbiotic processes exist in nature, some of them are very famous like the symbiosis between clownfish (like Nemo from the film) with sea anemone or for example lichens which consist in a symbiotic relationship between a fungus and a photosynthetic partner, usually a green alga.
In the plant kingdom we can find several types of symbiosis like the one between some ant species and certain acacia trees that protect these trees from harmful insects. Very specialized pollination processes are sometimes also considered as symbiotic relationships, since insects help the plant in their reproductive cycle and in turn insects get food from the flowers nectar.
The most studied symbiosis are beneficial plant microbe interactions, and among these the following two:
1) Legume-rhizobia symbiosis. This is a symbiosis between legumes (Fabaceae) which include many economically important crops like alfalfa, pea, soybean, peanut, etc… and soil bacteria collectively known as rhizobia. This symbiotic process leads to the formation of a new organ, the nodules, where the bacteria are capable of transforming the atmospheric nitrogen (N2) into NH4+ which can then be used by the plant, in a process known as biological nitrogen fixation. In turn, bacteria benefit from sugars coming from the photosynthesis. Biological nitrogen fixation is the key entry point of molecular nitrogen into the biogeochemical cycle of nitrogen.
2) Mycorrhizal symbiosis. This relationship represents the most widely spread plant symbiotic process1. Up to 90% of plant species are estimated to be able to be mycorrhized. This type of symbiosis consists in root colonization by different types of fungi. The plant thanks to the photosynthetic process provides the fungus with carbohydrates and in return the fungus substantially improves plant water and nutrient uptake capacity. Mycorrhizae can be ectotrophic (the fungi never enters in the plant cells) or arbuscular mycorrhyza, where the fungi hyphae penetrate in the plant cells.
For the initiation of a symbiotic process a signal exchange is needed between the two partners, to identify each other as a friend and not as an enemy. In 1990 it was described by Lerouge and collaborators2 that flavonoids secreted by legume roots induced in rhizobia the synthesis and secretion of a diffusible essential molecule needed for the symbiotic establishment. These symbiotic signals, named as Nod factors, consist in lipo-chitooligosaccharides (LCOs)3. Nod factors are essential for the specific recognition by host roots of the rhizobia and for the subsequent infection.
Mycorrhizal symbiosis is extremely ancient; it appeared more than 400 million years ago and seems to have accompanied the plant colonization of the terrestrial environment, what makes it so widely spread throughout the plant kingdom. The rhizobium-legume symbiosis apperared about 60 million years ago. Thus, it is probable that rhizobium-legume signaling mechanisms evolved from the AM symbiosis. Because of this and knowing that some legume genetic components are necessary both for nodulation and for mycorrhization, the same research group that in 1990 identified the Nod factors hypothesized that signals from the fungi could be similar to Nod factors. With this hypothesis, they used used as model the arbuscular endosymbiotic fungi Glomus intraradices and the legume Medicago truncatula to try to identify a putative mycorrhizal signal.
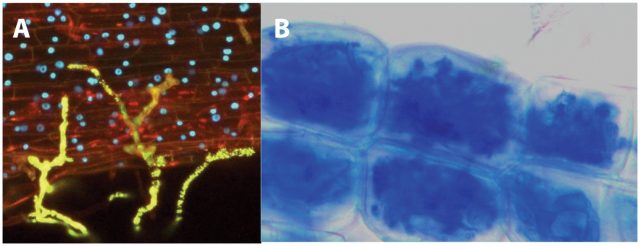
This french research group published in 2011, more than twenty years after the discovery of the nod factors, the identification of the structure of mycorrhizal signals 4. These molecules were named Myc factors of Myc-LCOs. To achieve this, the authors recovered the exudates from mycorrhized carrot roots and purified them using several biochemical methods, which included reverse-phase high-performance liquid chromatography (HPLC) followed by ultraperformance liquid chromatography coupled with quadrupole time-of-flight mass spectrometry (UPLC7Q-Tof MS). Interestingly, they were able to identifiy molecules corresponding to LCOs. The nature of these LCOs was confirmed by the use of LC and a 4000 Q-Trap mass spectrometer. These molecules are produced in such low amounts that to purifiy them it was necessary to extract three hundred litres of sterile exudates from mycrorrhyzed roots and exudates from 40 million germination spores of G. intraradices. Myc-LCOswere present in exudates of mycorrhized roots whilst in exudates of non-mycorrhized roots, grown in the same conditions but without fungi, no LCOs could be detected.

It was also obseverd that Myc-LCOs structure is remarkably similar to Nod factors as both signals have he same basic LCO structure of a chitin tetramer or pentamer (polymer of N-acetyl glucosamine) in which the N-acetyl group of the terminal non-reducing sugar is replaced by an acyl group. Briefly, the structure of the Myc-LCOs is simpler than Nod factors, having a limited number of chemical substitutions which can be related to the broad host range of mycorrhiza. Among others, one of the main differences observed between Myc-LCOs and Nod factors is that mycorrhizae produce LCOs with an oleic acid chain whereas no rhizobia strain has been shown to produce Nod factors with this structure.
Several experiments demonstrated that Myc-LCOs are able not only to stimulate the formation of mycorrhizal symbiosis but also to stimulate root branching, suggesting that, in the course of evolution, mycorrhizal simbyosis has selected fungal signals that can modify root development to facilitate further symbiotic infection.
To conclude, the identification of Myc-LCOs twenty years after Nod factors discovery represents a significant breakthrough in plant-microbe symbiotic interactions. Future genetic studies with fungi will surely help to advance in the evaluation of the importance of these molecules and in the knowledge of how a mycorrhizal symbiosis is established. Moreover, the discovery of Myc-LCOs also opens the door of exploring their use in agriculture practices to improve crops nutrient uptake.
References
- Corradi N, Bonfante P (2012) The Arbuscular Mycorrhizal Symbiosis: Origin and Evolution of a Beneficial Plant Infection. PLoS Pathog 8(4): e1002600.doi:10.1371/journal.ppat.1002600 ↩
- Lerouge P, Roche P, Faucher C, Maillet F, Truchet G, Promé JC, and Dénarié J (1990) Symbiotic host-specificity of Rhizobium meliloti is determined by a sulphated and acylated glucosamine oligosaccharide signal. Nature 344:781-784. ↩
- Gough C and Cullimore J (2011) Lipo-chitooligosaccharide signaling in endosymbiotic plant-microbe interactions. Mol Plant Microbe Interact. 24: 867–878. ↩
- Maillet F, Poinsot V, André O, Puech-Pagès V, Haouy A, Gueunier M, Cromer L, Giraudet D, Formey D, Niebel A, Martinez EA, Driguez H, Bécard G and Dénarié J (2011) Fungal lipochitooligosaccharide symbiotic signals in arbuscular mycorrhiza. Nature 469: 58-63. ↩
4 comments
Nice article Daniel! It’s a pleasure to find the topic of my research here. I’d been working on mycorrhizal fungi (and plants, obviously) for many years and I know that this kind of symbiosis is quite unknown for a lot of people.
I’m glad you have written about this.
“Thus, it is probable that rhizobium-legume signaling mechanisms evolved from the AM symbiosis.”
What is “AM symbiosis”? I think AM stands for “arbuscular mycorrhiza” but the acronym hasn’t been defined in the text…
You are right DarkSapiens, AM refers to arbuscular mycorrhiza.
Thanks for your comment Amara. I´m happy you liked the article