What does a molecular living fossil look like?
The idea of a living fossil is rather intuitive. We think that a kind of organism deserves that denomination if it has been living in the planet during a very large period of time apparently without any change. A well known example of living fossil is the Ginkgo tree (Ginkgo biloba) because, although it is currently the only extant member of a particular Gymnosperm clade, there are fossil Ginkgoaceae since the Jurassic. Other rather celebrated case is the coelacanth (Latimeria spp.), a lobe-finned fish group well known from the fossil record among paleontologists but only discovered as an extant living organism in the 20th century.
The concept of living fossil is rather tricky, since it is linked to our own perception. Fossil ginkgoes look similar to modern ones, and the whole fossil record of coelacanths seems pretty uniform since the Devonian, but are they really unchanged? Had Mesozoic ginkgos the same ecological preferences than modern ones? Were Permian coelacanths inhabitants of the ocean depths as they are today? Probably not. While some particular traits of a lineage may remain static through time, others may be changing faster. The bottom line is that we have an expectation from evolution: the longer a lineage has been outside there, the more changes we expect to find. If we think about it, it is the absence of change through long periods of geologic time (evolutionary stasis) what may seem rather intriguing itself.
One may wonder if living fossils can also be found in veiled traits of organisms that are not directly perceived. Can we find a living fossil in a molecular structure, such as a genome? A recent study published in Molecular Biology and Evolution1 may provide an example of such a case: the mitochondrial genome of mosses, whose structure has remained frozen since the Carboniferous period.
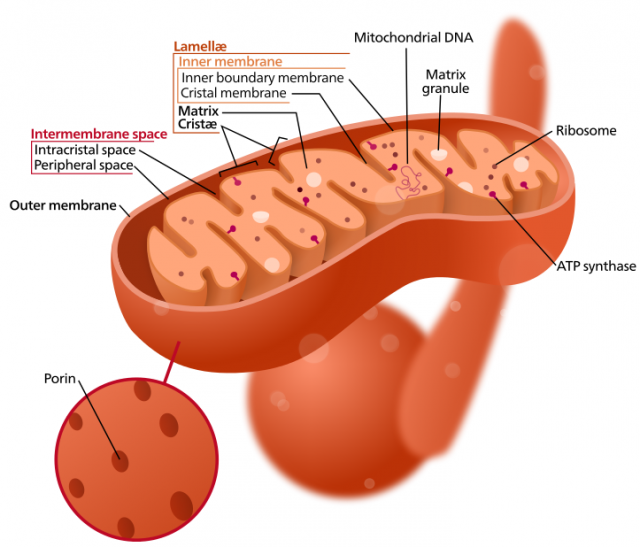
The mitochondrial genome, or chondrome, is a circular DNA molecule that contains the genes for many of the enzymes that are responsible of the metabolic reactions that take place in this organelle. Since we reached the genomic era, much research has been focused on the complete sequencing of organellar genomes, becoming a tool for the reconstruction of genomic evolution. Within plants, for instance, more than 70 mitochondrial genomes of different species and subspecies have been already sequenced. The comparative study of these genomes showed that this small DNA molecule is extraordinarily plastic in flowering plants. Its size varies greatly, and the gene order along the molecule changes very frequently, like if genes had a lot of freedom to switch positions (genomic rearrangements) among them. This activity is so frenetic that even within species of the same genus or even subspecies of the same species (lineages that diverged in relatively recent times) the gene order is not necessarily maintained.
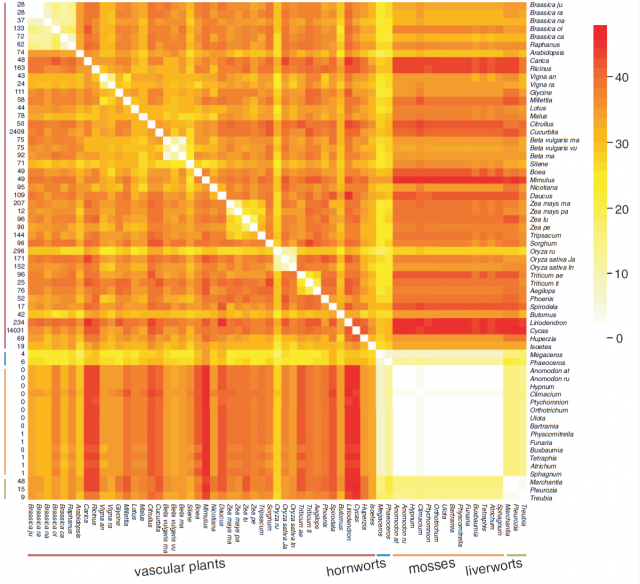
In striking contrast with flowering plants, the recent sequencing, assemblage and study of the mitochondrial genomes from 14 species of mosses showed a completely different viewpoint. Moss mitochondrial genomes are not only smaller than those of other plant lineages; they are also totally collinear: the gene order is exactly the same in all the 14 species examined.
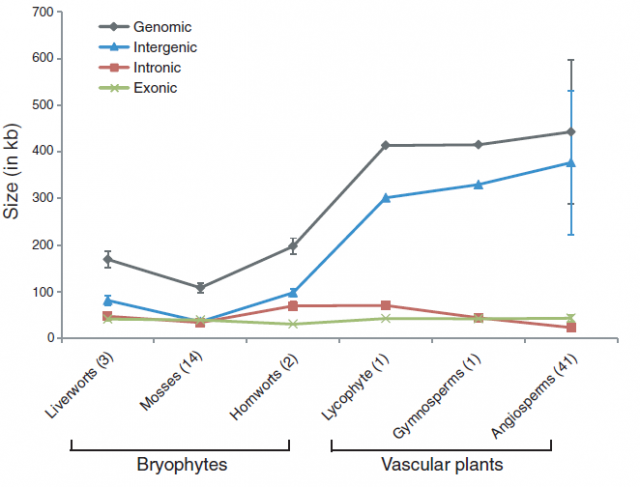
Just as with ginkgoes and coelacanths, it may be tempting (but wrong) to draw some general conclusions about moss evolution based on these facts. Does the stability of the chondrome mean that mosses have not evolved or that they are living fossils? Not at all! Mosses are a diverse group of plants hosting over 12,000 described species (more than other plant divisions such as ferns or gymnosperms) that had an active evolutionary history with extinctions and radiations, like other branches of the tree of life. However, during the time that this group of plants diversified and evolved, gaining new traits and losing others, the gene order in their mitochondrial DNA remained unchanged. The question is, why?
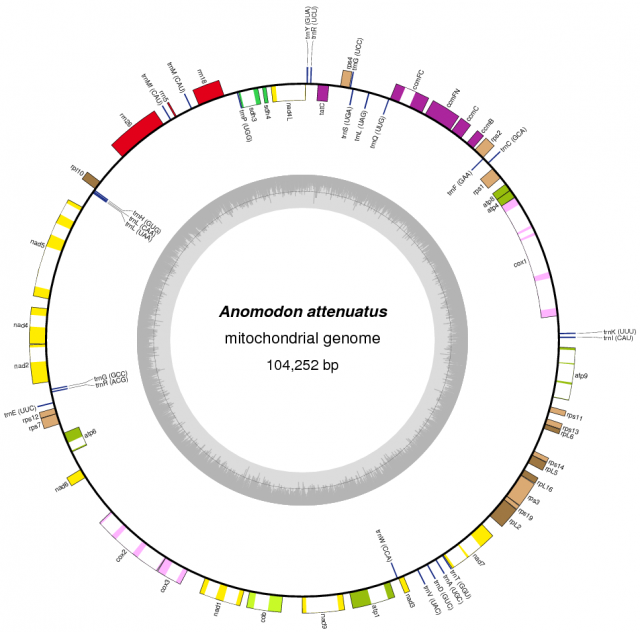
The causes of this stasis are not known, but one plausible explanation is related to the absence of repeated sequences in the moss chondrome. Repeated sequences are a requirement (but not a guarantee) for homologous recombinations that may result in gene rearrangements, and they are virtually absent in moss mitochondrial genomes, while they are much more frequent in vascular plants or other groups, such as a green algae.
Even if we assume that flowering plants have a particularly active mitochondrial genome, genomic rearrangements should be expected to occur given enough time in other divisions. This is why the stasis detected within mosses is intriguing: according to the available evidence, the same mitochondrial gene order has been maintained since the common ancestor of all known mosses evolved, 350 million years ago. This is, up to date, the longest period of mitochondrial genome stasis demonstrated in a whole plant lineage, and arguably a case of a molecular living fossil.
References
- Liu Y., Medina R. & Goffinet B. (2014). 350 My of Mitochondrial Genome Stasis in Mosses, an Early Land Plant Lineage., Molecular biology and evolution, PMID: http://www.ncbi.nlm.nih.gov/pubmed/24980738 ↩
1 comment
[…] mitokondrialen kasua, hauen egitura aldatu gabe mantentzen baita Karboniferotik. Rafael Medinaren What does a molecular living fossil look like? artikuluan datu […]