From a single progenitor to a pandemic multidrug-resistant Escherichia coli
From a single progenitor to a pandemic multidrug-resistant Escherichia coli
Few weeks ago, a new report by WHO 1 reveals that antibiotic resistance is now a major threat to global public health: “the world is headed for a post-antibiotic era”. One of the priorities is the treatment of urinary tract infections caused by Escherichia coli strains isolated from several countries and which are resistant to the third generation of cephalosporins and fluoroquinolones. Now, a recent paper published in PNAS 2 confirms the global dispersal of a single multidrug-resistant E. coli clone and demonstrated the role of mobile genetic elements and recombination in the evolution of this important pathogen.
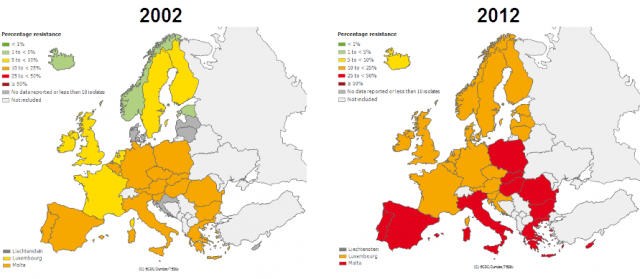
in EU during the last ten years. | Credit: European Centre for Disease Prevention and Control (ECDC).
E. coli ST131 (serotype O25:H4) is associated with human urinary tract and bloodstream infections and is also an example of globally disseminated multidrug-resistant bacteria. This strain was identified in 2008 as a major clone linked to the spread of extended-spectrum beta-lactamase CTX-M-15, a type of enzyme that provides resistance to beta-lactam antibiotics like penicillins, cephalosporins and derivatives. Since then, E. coli ST131 strains resistant to fluoroquinolones, aminoglycosides, trimethoprim-sulfamethoxazole and carbapenems have been reported, limiting treatment options for this pathogen: E. coli ST131 strains have become untreatable with standard antibiotics. The rapid emergence and successful spread of E. coli ST131 is associated with multidrug-resistance but also with several genes encoding virulence factor commonly associated with this kind of extraintestinal pathogenic E. coli (ExPEC).
The authors have used a global collection of 95 E. coli ST131 strains from six distinct geographical regions across the world (Australia, Canada, India, Spain, United Kingdom and New Zealand). Strains were isolated from clinical samples from 2000 to 2011. The strains were selected to ensure diversity with respect to geographic origin, date of isolation and clinical source. E. coli ST131 isolates were resistant to several antibiotics as aminoglycosides, penicillins, cephalosporins, fluoroquinolones, and sulphonamides. All genome strains were sequencing and phylogenetic analysis was carried out using whole genome alignment and single-nucleotide polymorphism analysis. All ST131 strains belonged to the same phylogroup and were distinct from other ExPEC, and three closely related clades were identified. Recombination events and mobile genetic elements have played a significant role driving the diversity within each clade. The results demonstrated that ST131 has arisen from a single progenitor E. coli that diverged into three sublineages some time before the year 2000, with acquisition of multiple mobile genetic elements, and associated recombination and point-mutations events.
The rapid global disseminations of E. coli ST131, combined with its multidrug-resistance and the lack of new antimicrobial drugs, highlights the urgent need to understand the evolution of this bacteria and combat its spread. E. coli ST131 is a good example of how a pandemic multidrug-resistance bacteria have evolved from a single clone and represents an important new public health threat.
References
- WHO’s first global report on antibiotic resistance reveals serious, worldwide threat to public health (30 April 2014) ↩
- Petty N.K., Ben Zakour N.L., Stanton-Cook M., Skippington E., Totsika M., Forde B.M., Phan M.D., Gomes Moriel D., Peters K.M. & Davies M. & (2014). Global dissemination of a multidrug resistant Escherichia coli clone., Proceedings of the National Academy of Sciences of the United States of America, PMID: 24706808 ↩
2 comments
[…] Hainbat botiken aurrean erresistentea den Escherichia coliren klon baten dispertsioa baieztatu da mundu osoan zehar. Honekin batera, agerian jarri da ere, elementu genetiko mugikorrek eta birkonbinazioek patogeno garrantzitsu honen eboluzioan izan duten papera. Ignacio López Goñik azaltzen digu From […]
[…] Se ha confirmado la dispersión por todo el mundo de un clon de Escherichia coli resistente a multitud de fármacos. También se ha puesto de manifiesto el papel que los elementos genéticos móviles y la recombinación han jugado en […]