From protein design to materials design
A key feature that makes natural materials highly sustainable and recyclable is their modularity. Biodegradable materials are such because biological organisms can digest and break their constituting chemicals into simpler building blocks. Often such simpler building blocks are then reused for energy or structural purposes. In other words, nature needs materials that can be broken into modular units, which can then be repurposed. Hence, looking at nature’s options to design biodegradable materials optimally is an obvious course of action.
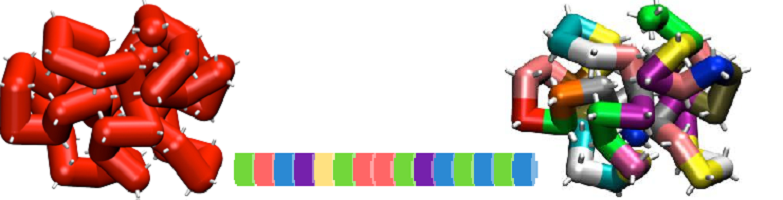
Among all materials found in nature, proteins are probably one of the most remarkable ones. They are the workhorse of life. They are the building infrastructure of living systems, the most efficient molecular machines known, and their enzymatic activity is still unmatched in versatility by any artificial system. Perhaps, proteins’ most remarkable feature is their modularity. The large amount of information required to specify each protein’s function is analogically encoded with an alphabet of just ~20 letters. All species use the same letters. Incidentally, that is why we eat other living organisms to find the materials we need for our bodies. The code consists of a simple string of amino acids assembled into a chain and connected by what is known as the peptide bond. Once the string is assembled, the proteins spontaneously perform their task like tiny machines. In fact, due to their scale, proteins are nature’s nano-machines.
How does an organism know the correct sequence to assemble for the needed function? Well, that information is written in the DNA and results from millions of years of selection through Darwinian evolution. It is this encoding process that allows for evolution in the first place because the latter operates by affecting the DNA code during the reproduction process. Still, it is the direct relationship between the code and the function that ultimately allows evolution to create new solutions. Hence, life could start to emerge from a chaotic mixture of chemicals only when a necessary function could be encoded into a string of monomers. From there, life gradually could explore new solutions and reach the explosion of variety that we observe today on our planet.
From all these considerations, we can deduce that a materials industry based on modular strings would be able to exploit all the benefits of natural materials and evolve towards higher versatility and sustainability.
Understanding how to convert a string into a function is a holy grail for material science. At the same time, this encoding process is a central biophysical problem in understanding how life could emerge.
In proteins, the study of how the function is encoded in a string is called protein design, while the prediction of the function of a given string is called protein folding.
If you want to know more, you can read a recently published review 1, where we go through the last 30 years of research to summarize the state of the art and highlight some applications related to fundamental protein evolution and design problems.
The review can be summarized into a critical finding. We discovered that directional interactions are enough for a material to allow the encoding of a function into a string using a limited set of monomers. Proteins use 20 amino acids, but it is known that protein can be built using only 4 or 5 monomers 2. If such directional interactions are present in entirely artificial polymers, we showed 345 that they can be designed or evolved like proteins to have desired target functions. That is why we call such polymers bionic proteins.
Such a result is not only beneficial for the design of new modular materials, but also for our constant search for life on other planets. The latter is because information encoding is the first stepping stone for Darwinian evolution to emerge. Hence, if the encoding is possible, it is not inconceivable that life could start from different materials and solvents other than protein in water, as it happened on Earth 6.
We classified [3–5] conventional synthetic polymers according to their potential capacity to encode functions as protein do. For instance, we identified polyurea, polyamide and polyurethane as possible candidates. Could you imagine a life was based on plastics?
Authors
Marcos Lequerica Mateos1, Irene Carbajo De La Guerra1, Ivan Coluzza1,2
1BCMaterials, Basque Center for Materials, Applications and Nanostructures, Bld. Martina Casiano, UPV/EHU Science Park, Barrio Sarriena s/n, 48940 Leioa, Spain
2Basque Foundation for Science, IKERBASQUE, 48009, Bilbao, Spain.
References
- Magi Meconi G, Sasselli I, Bianco V, Onuchic J, Coluzza I. Key aspects of the past 30 Years of protein design. Reports Prog Phys. 2022 Jun 15;85(8):86601. doi: 10.1088/1361-6633/ac78ef ↩
- Nerattini F, Tubiana L, Cardelli C, Bianco V, Dellago C, Coluzza I. Protein design under competing conditions for the availability of amino acids. Sci Rep . 2020 Dec 14;10(1):2684. doi: 10.1038/s41598-020-59401-9 ↩
- Cardelli C, Tubiana L, Bianco V, Nerattini F, Dellago C, Coluzza I. Heteropolymer Design and Folding of Arbitrary Topologies Reveals an Unexpected Role of Alphabet Size on the Knot Population. Macromolecules. 2018 Nov 13;51(21):8346–56. doi: 10.1021/acs.macromol.8b01359 ↩
- Cardelli C, Nerattini F, Tubiana L, Bianco V, Dellago C, Sciortino F, et al. General Methodology to Identify the Minimum Alphabet Size for Heteropolymer Design. Adv Theory Simulations 2019 Jul 7;2(7):1900031. doi: 10.1002/adts.201900031 ↩
- Coluzza I, van Oostrum PDJ, Capone B, Reimhult E, Dellago C. Sequence controlled self-knotting colloidal patchy polymers. Phys Rev Lett 2013 Feb 11 ;110(7):075501. doi: 10.1103/PhysRevLett.110.075501 ↩
- Lutz JF. Can Life Emerge from Synthetic Polymers? Isr J Chem. 2020;60(1–2):151–9. doi: 10.1002/ijch.201900110 ↩