CRISPRs, old sentinels for a new war

Under the threat of those annoying genetic parasites called viruses, eukaryotic cells developed, on the top of a wide innate defence, a curious and efficient RNA-based adaptive immune system referred as RNA interference or iRNA. You will find more info about this surveillance system, which detects and destroys foreign RNA molecules entering the cell with evil intentions, in this very house. However, after many years thinking that prokaryotes defend against viruses and other invasive nucleic acids using a variety of innate immune systems (e.g. DNA restriction), a totally different, analogous but not homologous, RNA-based adaptive immune system was found in them. Therefore, Bacteria and Archaea also have a system that provides them with “memory” of past aggressions as training for future attacks.
They began to draw the shape of this old sentinel in the late eighties, when sequencing techniques and bioinformatics were not even a shadow of what they are today. Ishino et al. cloned the gene of alkaline phosphatase isozyme from Escherichia coli and, after sequencing, they realized of a set of 29-nucleotide repeats separated by non-repetitive short sequences downstream of that gen. Afterwards, many more of these mysterious sequences were uncovered either in works addressing single gene research; or in whole genome studies of Bacteria an Achaea pushed by the sequencing technology that rose with the human genome project in 1990. Whatever it was the name of those sets of sequence repeats before, Jansen et al. in 2002 called them CRISPR (clustered, regularly interspaced short palindromic repeats), making reference to the peculiar structure of these regions within prokaryotic genomes (Fig.2A).
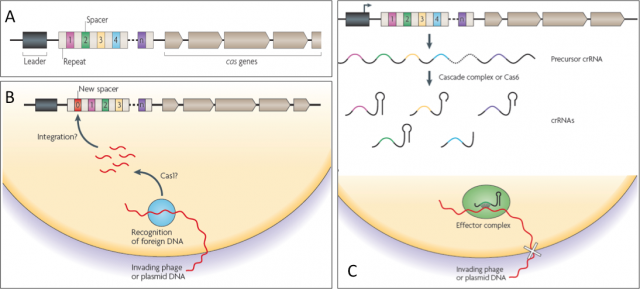
Given the variety in organization, conservation and type of Cas (CRISPR-associated protein) components found in these CRISPR systems, they have been sorted into three groups or types. They also have differences in the acquisition of new repeat-spacer units, in the CRISPR RNA biogenesis and in the interference mechanisms. Delve into detail on how they work is beyond the scope in this text, but all three share a common schema of action that many cases is not fully understood at molecular level yet (Fig. 2B and C). During a first encounter with an attacker, this sentinel learns the hallmarks of the enemy and makes sure to remember them well. When a stranger DNA (virus or plasmid) with dubious intentions invades the cell, the CRISPR dices it and inserts fragments generating new spacers within CRISPR locus. That will serve as memory and whenever the old enemy strikes again, the CRISPR locus will be transcribed in to a precursor RNA that, after being processed, will gide a effector complex to find the invader by sequence complementariety and destroying it by cutting with a nuclease activity.
In the last decade there has been a significant effort to elucidate the detailed molecular mechanism of these ancient prokaryotic sentinels. Glancing over, it just looks like another fancy system developed by the living matter to fend against the rigors of natural selection. A more careful look quickly considers number of details that makes it especially interesting, not only by its own nature but by its enormous potential. And this enormous potential is summarized in two of its main features. On the one hand, many CRISPR loci appear to encode all components (guide sequences togeter with processing and effector proteins) in relatively small DNA segments that can be handled well with current cloning technology; enabling their use in other organisms of interest. On the other hand, one can see that the cutting guide mechanism of the CRISPR effector complex is a nucleic acid, which currently can be modified at will, opening a range of possibilities for genetic engineering.
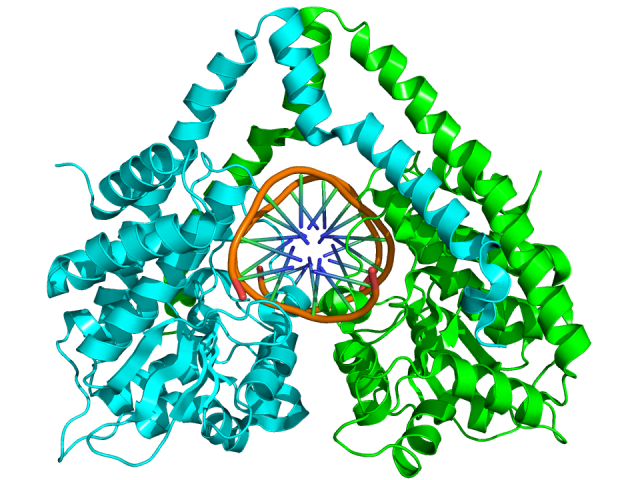
They say that, in the laboratories of biochemistry and genetic, these CRISPR nucleic acid guided nucleases could overtake the revered restriction enzymes long used to manipulate DNA. The later ones cut always on the same sequence guided by electrostatic interactions between its own peptidic sequence and the nucleic acid. We could say that their target is defined in their molecular structure (Fig. 3). Therefore, to change their target depends on modify the molecular structure of a protein, which is rather complex and not always possible. However, the nucleases from the CRISPR system will cut a target defined by the RNA generated in the CRISPR locus and that RNA, eventually, can be engendered in a laboratory.
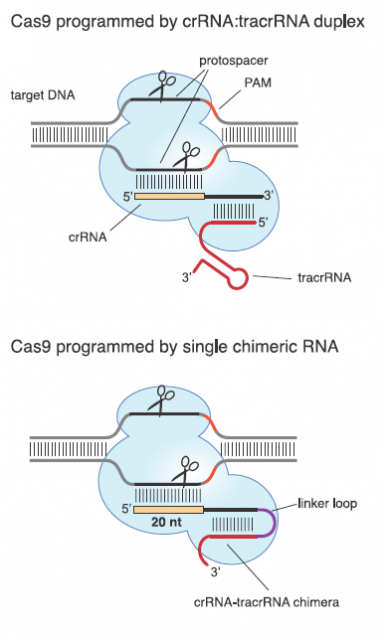
Those two main features suit them as valuable tool for molecular biology laboratories that copy, cut and paste DNA sequences in a daily bases. A very good proof of principal of this potential was provided in 2012 by a team led by Emmanuelle Charpentier1 They uncover the details and peculiarities in the molecular mechanism of interference for a subclass of CRISPR systems that uses Cas9 as nuclease. Afterwards they showed that Cas9 nuclease can be reprogramed by changing the DNA-binging sequence in an engineered RNA (Fig. 4).
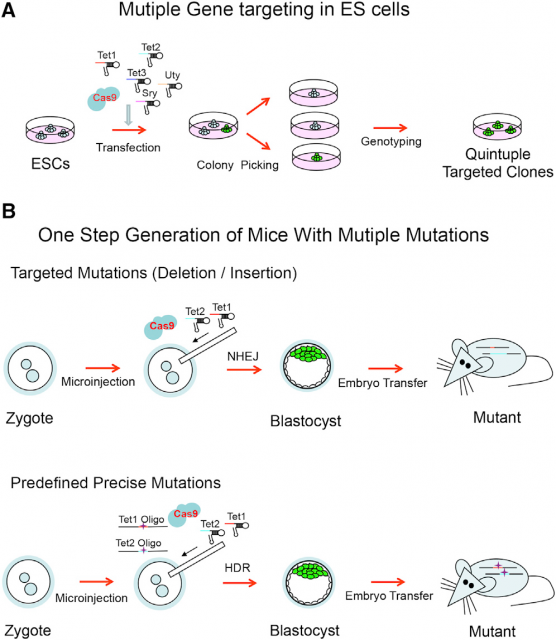
Going further, this old sentinel also raises a technology that in principle would be able to edit surgically the genome of any living being. In fact, several works have already shown that this is possible even with simultaneous editing of several genome sites in eukaryotic cells2 and mice3. Basically, what is done is transfect or inject this CRISPR/Cas9 system in cells in culture or in mouse embryos. Then, Cas9 will use the multiple guide sequences encoded into a single CRISPR array in a plasmid to cut specifically its DNA targets. Afterwards, the Non-homologous end joining (NHEJ) pathway that repairs DNA double-strand breaks (http://en.wikipedia.org/wiki/Non-homologous_end_joining/) will repair the nick in the sequence by a mutation prone mechanism due to the lack of DNA template to copy. This will usually insert a random mutation in the target sequence of the CRISPR/Cas9 system. However, if in the transfection/ injection process is included a donor plasmid or oligonucleotide with homology to the sequence flanking the DNA double-strand break, high-fidelity homologous recombination (http://en.wikipedia.org/wiki/Homologous_recombination/) will occur, integrating the new sequence in the genome of the organism (Fig. 5).
Apart from the improvement that CRISPR systems could bring to genetic research, also in undreamed ways, for example looping the loop about in CRISPR starts gettting recognition using them as RNA-guided transcription factors; no one ever look that his ability facilitating genetic editing in living organisms could be one of the pathways for gene therapy. Which, today is the hope for many diseases that plague us. However, it is still too early to talk about safety without side deletereous effects using CRISPR based system as in higher eukaryotes. Meanwhile, they are also taking advantage of features of this old sentinel to implement phage-resistant strains of industrial fermenting bacteria to avoid their masive dead due to viral infections. Therefore, though the first steps are promising and the potential of this system is high for several areas of the applied knowledge, much remains to be known and done before reaching humans. In any case, it is encouraging to see that, again, the old guards of our prokaryotic ancestors can be adapted to modern times to fight in our wars.
References
- Jinek, M. et al. (2012) A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science337, 816–21 ↩
- Cong, L. et al.(2013)Multiplex Genome Engineering Using CRISPR/Cas Systems. Science. 339(6121): 819–823 ↩
- Wang H., Chikdu S. Shivalila, Meelad M. Dawlaty, Albert W. Cheng, Feng Zhang & Rudolf Jaenisch (2013). One-Step Generation of Mice Carrying Mutations in Multiple Genes by CRISPR/Cas-Mediated Genome Engineering, Cell, 153 (4) 910-918. DOI: http://dx.doi.org/10.1016/j.cell.2013.04.025 ↩