Human Accelerated Regions: What makes us human?
Author: María Girbés Mínguez is a doctoral student at Center for Molecular Neurobiology Hamburg (ZMNH) / UKE (University Medical Center Hamburg-Eppendorf)
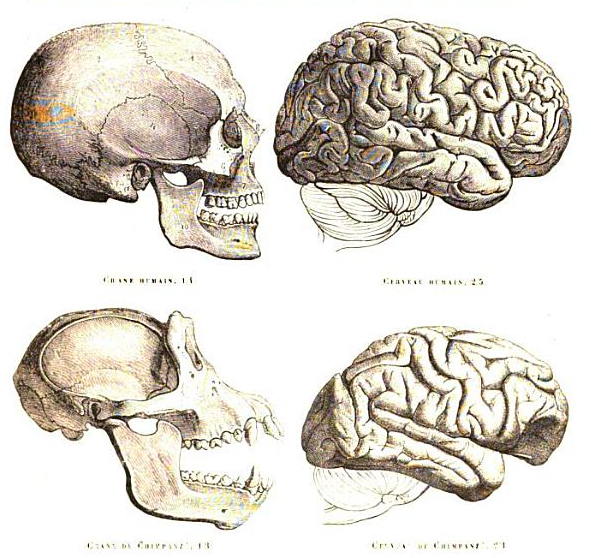
What differentiates us from our closest relatives and how are these differences caused? Scientists are trying to answer this question by comparing the genomic information of us and our closest relatives. Yet, we are far from having a genomic explanation that justifies the observed phenotypic differences.
Human Accelerated Regions (HARs)
However, some findings bring us closer to an answer. After the genome of the chimpanzee was published in 20052, Dr. Katherine S. Pollard and her team analysed this genomic data and compared it with the human genome. They developed a computer program to identify regions of the genome vastly different between chimpanzees and humans, but very conserved between chimpanzees and other vertebrate species. These regions would predict a loss or modification of function important for the generation of our species since we evolved from our common ancestor with chimpanzees. These sequences have come to be known as human accelerated regions (HARs)3.
The most surprising thing about HARs is that most of them are found in regions popularly known as “junk DNA”. These are DNA regions that do not encode for proteins, and most likely play a role in gene regulation. What could have made HARs be so conserved through the evolution of mammals, but be so different in humans? In fact the five most rapidly changed HARs have 26 times more substitutions in human when compared with chimpanzee, than the chimp HARs when compared with mouse, even though humans are more closely related to chimpanzees than chimpanzees are related to mice3.
Interestingly, most HARs (76%) appear to be positively selected for through evolution, meaning they turned out to be advantageous genetic variants which prevailed and were successful in the population4. Chimpanzees and humans diverged ~8 million years ago, and we still share about 99% of our genome1, suggesting that the genetic basis of the morphological differences must primarily stem from regions that regulate gene expression, with the potential of causing huge morphological changes5. Remarkably, HARs have been found located in intergenic regions and introns3, which could be regulatory sequences like enhancers or silencers of gene expression, implying that the differences present in the human HARs likely produce important changes in gene expression.
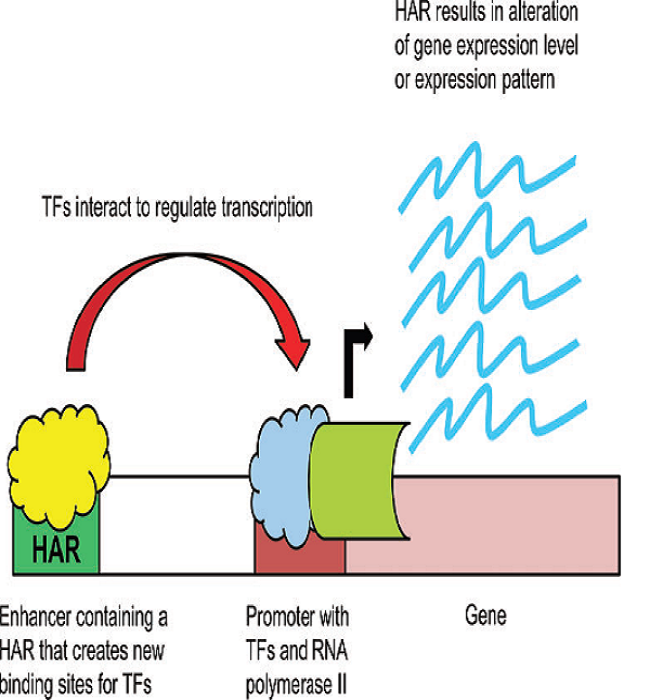
HARs in Archaic Humans
An interesting study aimed to quantify the degree of similarity between the HARs of modern humans and those of archaic members of the Homo genus (Neanderthals and Denisovans)6. Their study observed small differences (8.3%) between the HARs of archaic hominins and modern humans, meaning that most of the divergence must have taken place earlier than 500,000 years ago, and could have been a determinant event in the formation of the genus Homo.
However, around 10% the of mutations in HARs are polymorphic, meaning that a subset of people carry the mutated version, while others have the DNA sequence observed in chimpanzees7. These changes in HARs occurred recently in human evolution, and their occurrence indicates that they predate the migrations of humans around 60,000 years ago8.
HARs and brain disorders
Other lines of research aimed at investigating whether the mutated genes found in patients with schizophrenia are often located in close proximity to HARs, and therefore might be regulated by HARs9. Since schizophrenia is a brain disorder that impairs high mental functions characteristic of modern humans, the authors expected that genes or regulatory regions involved in the pathogenesis of schizophrenia would be exclusive to the human brain.
The authors found indeed that the genes mutated in schizophrenia patients were in close proximity to HARs. Since genes in proximity to HARs are likely to be under their regulation, this suggest important functional roles of HARs in the genetic architecture of schizophrenia. The authors did not stop here, but evaluated which of these schizophrenia associated genes in close proximity to HARs were in addition highly conserved in primates, and found that those genes were the most conserved. These results could seem counterintuitive, but they imply that HARs are regions that evolved divergently in the human lineage under positive selection, and that genes that are functionally important for humans gained new regulation of transcription thanks to HARs.
Furthermore, researchers investigating genetic variants present in patients with autism, found out that these genetic variants are often located in HARs10. In addition, these particular HARs seem to interact with proteins that regulate gene expression in the brain, and are of great importance for several intracellular processes during not only development, but also correct functioning in adulthood.
Discovering the functions of HARs
Researchers are exploring the possible functions of HARs, and the role they might have played in human evolution, but there is still a long way to go. Fortunately, recent discoveries in biotechnology are proving helpful, since it is possible to obtain different cell types from our closest relatives after obtaining skin cells (fibroblasts), reprogram them to become pluripotent stem cells (cells that can differentiate into different cell types), and edit their DNA to obtain modified brain, heart, or liver cells11. It is even possible to generate a model of the differentiation of the human cortex using human fibroblasts as a start12. These cells allow researchers to observe the changes in gene regulatory networks caused by HARs in the primate and human contexts, and the effects on neural development and functions.
These research directions may advance our understanding, and fill out the puzzle of what makes us human, and how the constraints of evolution shaped our nature.
References:
(1) Chimpanzee Sequencing and Analysis Consortium. Initial Sequence of the Chimpanzee Genome and Comparison with the Human Genome. Nature 2005, 437 (7055), 69–87. doi: 10.1038/nature04072.
(2) International Human Genome Sequencing Consortium. Initial Sequencing and Analysis of the Human Genome. Nature 2001, 409 (6822), 860–921. doi: 10.1038/35057062.
(3) Pollard, K. S.; Salama, S. R.; King, B.; Kern, A. D.; Dreszer, T.; Katzman, S.; Siepel, A.; Pedersen, J. S.; Bejerano, G.; Baertsch, R.; Rosenbloom, K. R.; Kent, J.; Haussler, D. Forces Shaping the Fastest Evolving Regions in the Human Genome. PLoS Genet 2006, 2 (10), e168. doi: 10.1371/journal.pgen.0020168.
(4) Kostka, D.; Hubisz, M. J.; Siepel, A.; Pollard, K. S. The Role of GC-Biased Gene Conversion in Shaping the Fastest Evolving Regions of the Human Genome. Molecular Biology and Evolution 2012, 29 (3), 1047–1057. doi: 10.1093/molbev/msr279.
(5) Levchenko, A.; Kanapin, A.; Samsonova, A.; Gainetdinov, R. R. Human Accelerated Regions and Other Human-Specific Sequence Variations in the Context of Evolution and Their Relevance for Brain Development. Genome Biology and Evolution 2018, 10 (1), 166–188. doi: 10.1093/gbe/evx240.
(6) Green, R. E.; Krause, J.; Briggs, A. W.; Maricic, T.; Stenzel, U.; Kircher, M.; Patterson, N.; Li, H.; Zhai, W.; Fritz, M. H. Y.; Hansen, N. F.; Durand, E. Y.; Malaspinas, A. S.; Jensen, J. D.; Marques-Bonet, T.; Alkan, C.; Prufer, K.; Meyer, M.; Burbano, H. A.; Good, J. M.; Schultz, R.; Aximu-Petri, A.; Butthof, A.; Hober, B.; Hoffner, B.; Siegemund, M.; Weihmann, A.; Nusbaum, C.; Lander, E. S.; Russ, C.; Novod, N.; Affourtit, J.; Egholm, M.; Verna, C.; Rudan, P.; Brajkovic, D.; Kucan, Z.; Gusic, I.; Doronichev, V. B.; Golovanova, L. V.; Lalueza-Fox, C.; de la Rasilla, M.; Fortea, J.; Rosas, A.; Schmitz, R. W.; Johnson, P. L. F.; Eichler, E. E.; Falush, D.; Birney, E.; Mullikin, J. C.; Slatkin, M.; Nielsen, R.; Kelso, J.; Lachmann, M.; Reich, D.; Paabo, S. A Draft Sequence of the Neandertal Genome. Science 2010, 328 (5979), 710–722. doi: 10.1126/science.1188021.
(7) Hubisz, M. J.; Pollard, K. S. Exploring the Genesis and Functions of Human Accelerated Regions Sheds Light on Their Role in Human Evolution. Current Opinion in Genetics & Development 2014, 29, 15–21. doi: 10.1016/j.gde.2014.07.005.
(8) Krause, J.; Pääbo, S. Genetic Time Travel. Genetics 2016, 203 (1), 9–12. doi: 10.1534/genetics.116.187856.
(9) Xu, K.; Schadt, E. E.; Pollard, K. S.; Roussos, P.; Dudley, J. T. Genomic and Network Patterns of Schizophrenia Genetic Variation in Human Evolutionary Accelerated Regions. Molecular Biology and Evolution 2015, 32 (5), 1148–1160. doi: 10.1093/molbev/msv031.
(10) Doan, R. N.; Bae, B.-I.; Cubelos, B.; Chang, C.; Hossain, A. A.; Al-Saad, S.; Mukaddes, N. M.; Oner, O.; Al-Saffar, M.; Balkhy, S.; Gascon, G. G.; Homozygosity Mapping Consortium for Autism; Nieto, M.; Walsh, C. A. Mutations in Human Accelerated Regions Disrupt Cognition and Social Behavior. Cell 2016, 167 (2), 341-354.e12. doi: 10.1016/j.cell.2016.08.071.
(11) Gallego Romero, I.; Pavlovic, B. J.; Hernando-Herraez, I.; Zhou, X.; Ward, M. C.; Banovich, N. E.; Kagan, C. L.; Burnett, J. E.; Huang, C. H.; Mitrano, A.; Chavarria, C. I.; Friedrich Ben-Nun, I.; Li, Y.; Sabatini, K.; Leonardo, T. R.; Parast, M.; Marques-Bonet, T.; Laurent, L. C.; Loring, J. F.; Gilad, Y. A Panel of Induced Pluripotent Stem Cells from Chimpanzees: A Resource for Comparative Functional Genomics. eLife 2015, 4, e07103. doi: 10.7554/eLife.07103.
(12) Espuny-Camacho, I.; Michelsen, K. A.; Gall, D.; Linaro, D.; Hasche, A.; Bonnefont, J.; Bali, C.; Orduz, D.; Bilheu, A.; Herpoel, A.; Lambert, N.; Gaspard, N.; Péron, S.; Schiffmann, S. N.; Giugliano, M.; Gaillard, A.; Vanderhaeghen, P. Pyramidal Neurons Derived from Human Pluripotent Stem Cells Integrate Efficiently into Mouse Brain Circuits In Vivo. Neuron 2013, 77 (3), 440–456. doi: 10.1016/j.neuron.2012.12.011.
2 comments
Fascinating, so close and so far, because expression…
[…] Zerk ezberdintzen gaitu gizakiak primateengandik? Oraindik ez da topatu erantzun genetikoa. Baina aurrerapenak egiten ari dira. María Gibésen Human Accelerated Regions: What makes us human? […]