STOP the STOP in cancer: evading pathogenic premature translation termination of tumor suppressors
Author: Rafael Pulido is an Ikerbasque Research Professor at the BioCruces Health Research Institute

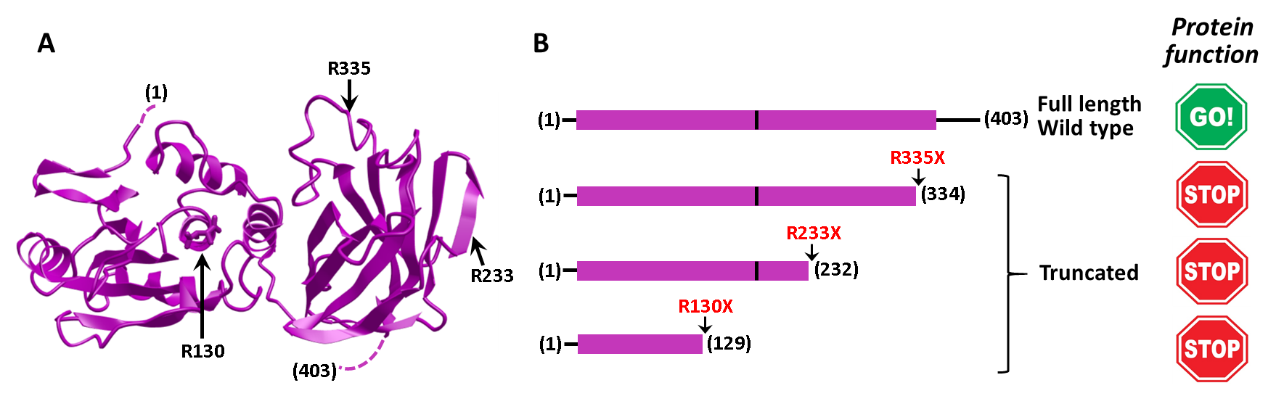
Inherited or somatic mutations targeting genes which encode essential cell growth regulatory proteins favor the emergence of cancer. Cells carrying these mutations acquire growing and survival advantages that trigger the emergence of the tumor. A large number of tumor-associated mutations, as well as many mutations found in the germline of hereditary cancer patients, are single-nucleotide nonsense substitutions that generate stop codons in coding regions (premature termination codons, PTC) of tumor suppressor genes. The biosynthetic translation of these genes carrying PTC mutations produces truncated unstable or non-functional tumor suppressor proteins that lack the ability to control cell growth. Nonsense substitutions targeting one of the tumor suppressor genes more mutated in cancer, PTEN, represent about the 20% of PTEN mutations found in tumors, and about 50% of patients with germline PTEN mutations and cancer predisposition (PTEN Hamartoma Tumor Syndrome patients) carry a nucleotide nonsense substitution generating a PTC. We have coined the term PTCome to refer to the collection of PTC that are found in a specific gene in association with disease. The PTEN PTCome is composed by more than 140 different PTC mutations, which distribute along almost the entire PTEN coding sequence. Three of these mutations are found with high prevalence, and together account for about 30% of the PTEN PTC mutations detected in patients. The truncated PTEN proteins generated by translation of these mutations are non-functional (Figure 1), and the restoration of full-length functional PTEN biosynthesis in these cases could help to compensate the pathogenicity of the mutation.
Translational readthrough of PTC consists in the incorporation of an amino acid, during protein biosynthetic translation, in the position corresponding to the anomalous PTC created by a nonsense mutation in the coding region of a gene. The readthrough process is facilitated by the weakening of the fidelity in the recognition of the PTC by the translation termination machinery, and can be further improved by compounds that bind to the ribosome, including some antibiotics currently used in the clinics (Figure 2). The efficiency of the induced readthrough depends on the inducer compound, as well as on the identity of the PTC (TAA, TAG, or TGA) and its surrounding nucleotide sequence. Importantly, compounds inducing PTC readthrough do not affect the efficient recognition of natural termination codons. The heterogeneity in the efficacy and toxicity of the readthrough-inducing compounds is one of the major restrictions of this approach for its therapeutic application. In addition, for proteins such as tumor suppressors it is also a limiting aspect their functional status after the readthrough reconstitution, since this process incorporates variable amino acids (encoded by near-cognate codons) at the PTC position. This demands dedicated studies that address the potential of precision anticancer therapies based on the functional reconstitution of tumor suppressors targeted by PTC.
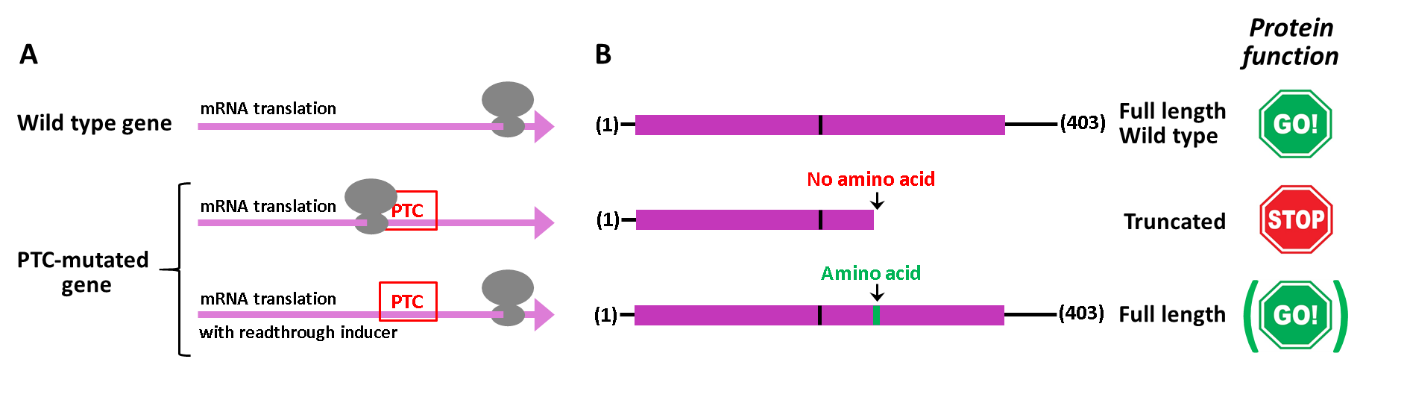
To gain knowledge on the potential of PTEN PTC readthrough as an anticancer therapy, we have performed a global biochemical and functional analysis of the readthrough of the entire PTEN PTCome. We have found a heterogeneous response of the different PTEN PTC mutations to readthrough induced by aminoglycoside antibiotics, ranging from 0 to 35% of biochemical full-length protein reconstitution, and we have determined the function of the resulting PTEN protein variants 1. Our analysis has unveiled the set of PTEN PTC mutations that can be functionally reconstituted by translational readthrough, among which the more prevalent mutations associated to disease (Figure 1) are included. The results from these in vitro experiments open the possibility of readthrough therapeutic actionability on specific groups of cancer patients with PTEN PTC mutations. Further evidence is necessary, using physiologic experimental conditions, to confirm the benefits of this approach to rescue PTEN functions in cancer patients.
Our global studies on the functional reconstitution by readthrough of the PTEN PTCome establish the basis of the preclinical experimental methodology required to implement with efficiency readthrough-based therapies in precision oncology. These global analyses should be extended to the PTCome from other important tumor suppressors, from which genetic information is starting to be routinely obtained in the clinic. This will make accessible useful information on the specific readthrough response expected for cancer cases affected by PTC mutations at relevant tumor suppressors. The identification and preclinical validation of PTC readthrough-inducing compounds of high efficacy and low toxicity will provide the tools that bring readthrough anti-cancer therapies to practice. The availability of the readthrough response of PTCome from tumor suppressors will accelerate the attempts to evade their pathogenic premature translation termination in human cancer.
References
- Luna, S., Torices, L., Mingo, J., Amo. L., Rodríguez-Escudero, I., Ruiz-Ibarlucea, P., Erramuzpe, A., Cortés, J.M., Tejada, M.I., Molina, M., Nunes-Xavier, C.E., López, J.I., Cid, V.J., Pulido, R. (2021). A global analysis of the reconstitution of PTEN function by translational readthrough of PTEN pathogenic premature termination codons. Hum. Mutat. doi: 10.1002/humu.24186; PMID: 33600059 ↩
1 comment
[…] Tumorazio ezabatzaileen sintesia gelditzen dute minbizia agertzea eragiten duten mutazioak, ingurumenekoak ala heredatutakoak. Gelditze hori gelditzea (stop the stop) minbiziari aurre egiteko bidea da. Rafael Pulidok Biocrucesen bere taldearekin: STOP the STOP in cancer: evading pathogenic premature translation termination […]