MitoEM challenge remains open
MitoEM challenge remains open
Mitochondria are the primary energy providers for cell activities. The quantification of the size and geometry of mitochondria is crucial to basic neuroscience research, as it contributes to the identification of neuron types. This quantification is also informative to clinical studies where energy-usage unbalances are implied, like bipolar disorder or diabetes. High-resolution imaging technologies like electron microscopy (EM) have been used to reveal the detailed 3D geometry of mitochondria at the nanometer level with terabyte data scale. Consequently, to enable an in-depth biological analysis, we need high-throughput and robust 3D mitochondria instance segmentation methods.
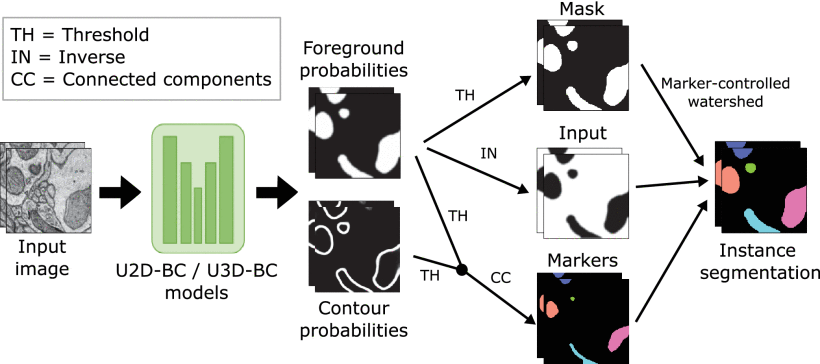
The Large-scale 3D Mitochondria Instance Segmentation challenge (MitoEM), presented at the IEEE International Symposium on Biomedical Imaging (ISBI) 2021, is the first open comparison of mitochondria instance segmentation algorithms on EM volumes. Now, a team of researchers analyzes 1 the current progress in the mitochondria segmentation task based on the results of MitoEM, and presents an in-depth analysis of the state-of-the-art evaluation metrics for identifying mitochondria instance segmentation errors. This analysis reveals the difficulties of the current approaches and can be used as a guide for the creation of the next generation mitochondria segmentation models.
The basis for the MitoEM challenge is a previously released large-scale 3D mitochondria instance segmentation benchmark, known as the MitoEM dataset. The MitoEM dataset comprises two 3D EM image stacks. These image stacks originate from distinct sources, one from adult rat brain tissue (MitoEM-R) and the other from adult human brain tissue (MitoEM-H). Notably, the MitoEM dataset represents a substantial increase in scale, being approximately 1,986 times larger than the previous de facto benchmark dataset, the EPFL Hippocampus dataset, the so-called Lucchi benchmark. From the 1,000 consecutive slices of each stack, ground-truth mitochondria instance labels were provided for the first 500 slices and split into training (400 slices) and validation (100 slices) subsets. The annotations of the remaining 500 slices of each volume were kept private and used as the test set.
The challenge was accepted to ISBI 2021 in October 2020 and officially announced in November 2020. This announcement was accompanied by the creation of a dedicated website and the preparation of an evaluation system. The two image volumes, MitoEM-R and MitoEM-H, were made immediately available to participants to enable them to begin developing their methods. Participants performed the segmentation on their own computers. The challenge was widely advertised and was open to any interested participants. A total of 257 individuals registered for the challenge and 14 teams submitted their results. Eight teams successfully completed the challenge.
Posterior to the challenge, annotation errors in the ground truth were corrected without altering the final ranking. The authors present in this new paper the eight top-performing approaches from the challenge participants, along with their own baseline strategies. But they also take a critical look at the challenge itself. A retrospective evaluation of the scoring system revealed that: 1) the challenge metric was permissive with the false positive predictions; and 2) the size-based grouping of instances did not correctly categorize mitochondria of interest.
Consequently, the researchers propose a new scoring system that better reflects the correctness of the segmentation results. Although several of the top methods are compared favourably to their own baselines, substantial errors remain unsolved for mitochondria with challenging morphologies. Thus, the challenge remains open for submission and automatic evaluation, with all volumes available for download.
Author: César Tomé López is a science writer and the editor of Mapping Ignorance
Disclaimer: Parts of this article may have been copied verbatim or almost verbatim from the referenced research paper/s.
References
- D. Franco-Barranco et al. (2023) Current Progress and Challenges in Large-Scale 3D Mitochondria Instance Segmentation IEEE Transactions on Medical Imaging doi: 10.1109/TMI.2023.3320497 ↩