Huisgenases, new protein catalysts which are not enzymes
Huisgenases, new protein catalysts which are not enzymes
Proteins can perform a huge number of biological functions with amazing efficiency. In order to achieve these different functions, proteins rely on the precise 3D arrangement of functional groups which are referred to as the protein fold. Some of these functions include acting as a catalyst in biochemical reactions; in these cases proteins are called enzymes.
The enzyme’s amino acid sequence determines its ability to fold into a defined native structure, that will be the responsible for the dynamics of its functional groups, required for binding to the substrate and, thus, allowing catalysis. Many efforts have been devoted to understand how proteins evolve to adopt new functions; the consensus emerged that new functions originate from latent promiscuous enzymes or broad specificity enzymes that serve as starting points for further evolution.
The artificial redesign of existing proteins in order to perform new reactions has been very fruitful. These approaches rely mostly on the grafting of active catalytic sites or catalytic cofactors onto protein scaffolds. Repeat proteins are of interest as scaffolds, as their repeated structures and the extrapolation from known structures can provide geometric parameters and simple sequence-to-structure relations to guide the rational design of new scaffolds. Additionally, they offer possibilities for systematic variation of repeated elements to produce different families of proteins.
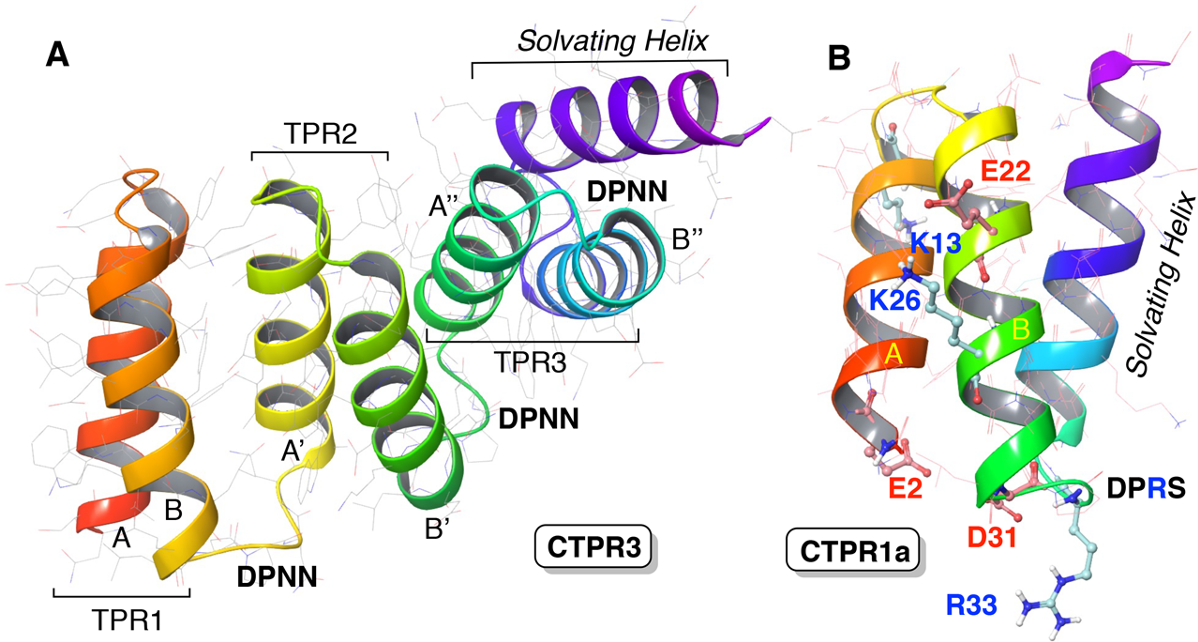
Now, a team of researchers has designed 1 repeat proteins able to catalyze the 1,3-dipolar reaction between an imine and a π-deficient dipolarophile in a tetrahydrofuran solution to form unnatural nitroproline esters, a reaction that no enzyme can catalyze.
Several groups have been successful already on different enzymatic activities using alpha helices as the principal structural component and combining multiple helical elements. Interestingly, helical based scaffolds led to catalytic activities through very different approaches. This new work takes this same direction: repeat proteins that fold in helix−turn−helix structures as biocatalysts for new reactions. The researchers have explored a minimalist approach, though, using consensus repeat proteins, which display acid and basic residues that can act as catalytic active sites for the Huisgen 1,3-dipolar reaction between imines and a π-deficient dipolarophile.
Enzymes can be classified in six different groups depending upon the type of reaction they catalyze: oxidoreductases, transferases, hydrolases, lyases, isomerases, and ligases. As a consequence of this new work a new category is proposed, Huisgenases, for proteins that catalyze this kind of thermal pericyclic reactions. No enzyme-assisted direct Huisgen 1,3-dipolar reaction has been observed so far, meaning that, even though Huisgenases are biomaterials, none have been identified in living systems.
The results can be a stimulus for a second generation of Huisgenases either with an optimized catalytic site or including more restricted transition metal-containing active sites, which could catalyze the diastereo- and enantioselective formation of unnatural proline amino acids, thus expanding the repertoire of chemical reactions promoted by synthetic enzymes.
Author: César Tomé López is a science writer and the editor of Mapping Ignorance
Disclaimer: Parts of this article are copied verbatim or almost verbatim from the referenced research paper.
References
- Iván Rivilla, Mikel Odriozola-Gimeno, Antonio Aires, Ana Gimeno, Jesús Jiménez-Barbero, Miquel Torrent-Sucarrat, Aitziber L. Cortajarena, and Fernando P. Cossío (2020) Discovering Biomolecules with Huisgenase Activity: Designed Repeat Proteins as Biocatalysts for (3 + 2) Cycloadditions JACS doi: 10.1021/jacs.9b06823 ↩