Loperamide and autism
Occasionally I receive groups of messages from families with autistic members and I know the reason: they have read in the press exciting news about autism, a spectacular treatment, even a miracle cure. Unfortunately, if all that news were true we would have solved autism hundreds of times by now. So let’s start with the harsh reality: autism has no cure and from what we know about its origin, a strong genetic component that generates alterations in brain circuits, it probably never will. This does not prevent us from doing a lot to improve the quality of life of autistic people and their families, to help them to improve, to have dignified and happy lives, lives worth living. And often it’s not up to them, it’s up to us. The latest news of this kind was that loperamide (under its different commercial names: Fortasec, Lopex, Regulane, Fadal, Imodium, Dimor, Loperam, Salvacoll, Lomotil, Antilax), a very common medication used for diarrhea could improve the symptoms of autism.
When you get a news item like this, the only sensible thing to do is to look for the original source. If it only appears in the media or on websites, it is a bad sign. All over the world, research is published in specialized journals after the article has been evaluated by experts in the same field, what we call peer review. Here the good news is that there was a scientific article, published by Elise Koch and Ditte Demontis in the journal Frontiers in Pharmacology 1. That’s another useful clue. The really important papers, a cure for autism would be the case, are usually published in three journals, Science, Nature and Cell. But it is also true that if the research is really groundbreaking, the researchers are little known or the study has some weak flank, it is possible that the authors will not get it published in these three “top” journals and will have to settle for a lower tier. Frontiers in Pharmacology is not in that select group of excellence, but it is not a bad journal either. It has an intermediate level and many Chinese research groups have been publishing in it lately, a sign of the times. The two researchers are Norwegian and Danish, two countries that are not world leaders in science, but where hundreds of research groups do honest and serious work.
Bringing a new drug to market is a process that costs hundreds or billions of euros and 10-15 years of time. That is why it was a spectacular success for science, particularly European science, to achieve an effective vaccine against the coronavirus in one year. Can you imagine 10-15 years with the pandemic raging? I doubt that our civilization could have withstood it. Prior studies, before going to market, cover two major aspects: that the drug is safe and that it is effective. A very interesting derivative arises here. Current drugs have shown that their consumption is safe, and it has also been seen that patients with a disease or condition, if they take it, improve more than if they take a placebo, a harmless substance without an active ingredient. But if we find a second disease where the drug is effective, it is very interesting because we do not have to do all the safety controls again, because they are already done. A second indication, being useful for another disease, is a bargain for all parties involved, because the costs are lower and the potential benefits increase. That is the origin of Koch and Demontis’ work.
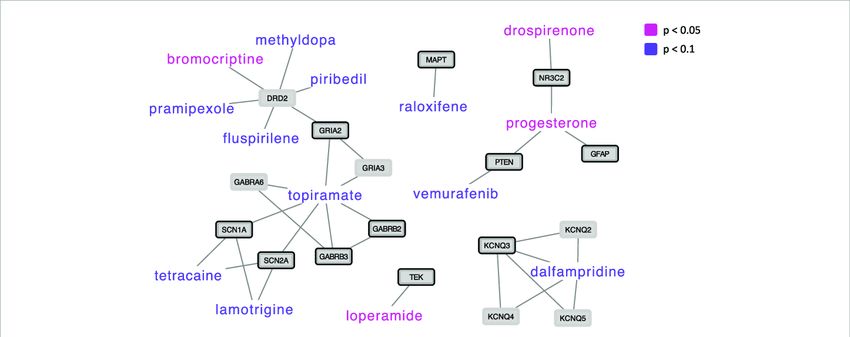
To look for possible interactions, the two Scandinavian researchers did the following. They started with the genes that supposedly increase the risk of having autism spectrum disorders (ASD) and studied the protein-protein interaction networks of the products of these genes. They then identified approved drugs known to interact with proteins within this network. They then assessed whether these drugs can normalize ASD-associated gene expression perturbations in ASD network genes. This was done by analyzing drug-induced versus ASD-associated gene expression, identifying drug-caused gene expression perturbations that were opposite to ASD-associated perturbations. Following this study, the two scientists identified four drugs that showed significant (p < 0.05) gene expression changes opposite to the changes observed in ASD: loperamide, bromocriptine, drospirenone and progesterone. Loperamide, as we mentioned, is one of the most widely used drugs against diarrhea. Finally, these drugs act on biological systems related to ASD, indicating that theoretically these drugs could effectively treat the core symptoms of ASD.
The most important thing is to understand what the researchers have and have not done. This has been the procedure:
1.- Choosing ASD genes
They used two different sources to define ASD genes: the latest large-scale GWAS on ASD, which included 18,381 individuals with ASD and 27,969 controls 2, and the largest ASD exome sequencing study (N = 35,584 total samples, 11,986 individuals with ASD) 3. 54 ASD-related genes were identified by GWAS and 102 ASD risk genes were identified in the whole exome study. Of these, 7 genes were already included from GWAS, resulting in 149 candidate ASD-risk genes in total. Of these 149 genes, 147 generate proteins present in the interactome, the map of molecular interactions between proteins.
2.- Using the human protein interactome
This interactome was constructed from data from 15 commonly used databases, focusing on high quality protein-protein interactions (PPIs). This interactome consists of 17,706 unique proteins (nodes) interconnected by 351,444 IPPs (edges or links), resulting in 346,330 IPPs after removing self-loops.
3.- Defining the ASD network
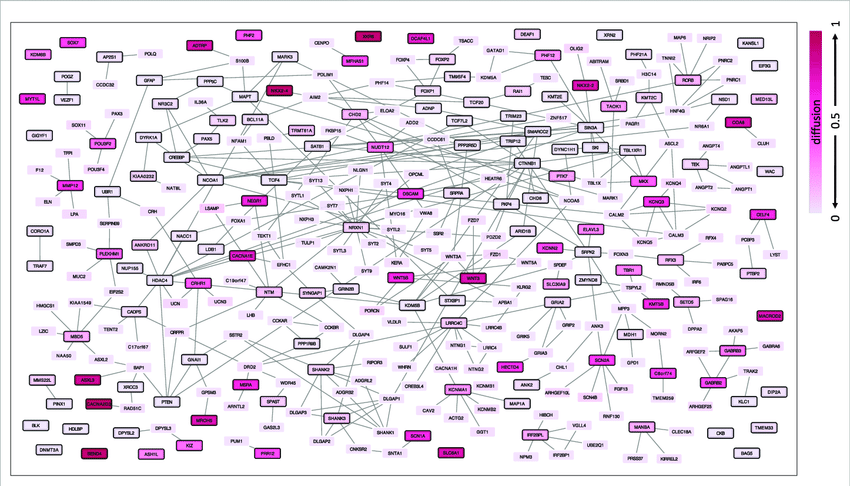
Most of the approved drugs do not target the proteins altered by the disease, but bind to neighboring proteins in the network. Therefore, the researchers defined an ASD network that includes not only the ASD genes described above, but also genes in their immediate network proximity. The network proximity between proteins is scored based on their IPPs, where higher diffusion output values are related to higher relatedness to input proteins. Genes defined as ASD genes were used as input query genes, and the top 1% of diffusion output proteins were included in the ASD network. The annotation categories of gene ontology molecular function, biological process, cellular component and pathways, and disease were included.
4.- Analyze the drug target network.
They used the drug-gene interaction database (DGIdb) to identify drug-gene interactions between approved drugs and genes in the ASD network. They studied 439 approved drugs.
5.- Gene expression perturbation profiles.
For drugs that interact with ASD network genes, they used gene expression data (drug vs. non-drug) to assess whether these drugs modulate the ASD of network genes. To assess whether drug repurposing candidates could change ASD-associated gene expression perturbations (whether they down-regulate ASD up-regulated genes or vice versa), Spearman’s ρ correlation between drug-induced perturbations and ASD-associated perturbations was calculated for each drug, where negative correlation coefficients indicate that the drug could reverse ASD-associated gene expression changes.
In summary, what have the two researchers NOT done? First, they have not, at least in this paper, worked with people with ASD. It is a whole process done on a computer, a bioinformatics study based on databases. Second, they have not given a loperamide pill to anyone. Today, there is no evidence in people or animals that this drug improves the symptoms of autism. What have they DONE? A study to identify approved drugs for their possible potential to treat ASD, and they have found four, a starting point, no more, no less.
References
- Koch E, Demontis D (2022) Drug repurposing candidates to treat core symptoms in autism spectrum disorder. Front Pharmacol 13: doi: 10.3389/fphar.2022.995439 ↩
- Grove J, Ripke S, Als TD, Mattheisen M, Walters RK, Won H, Pallesen J, Agerbo E, Andreassen OA, Anney R, Awashti S, Belliveau R, Bettella F, Buxbaum JD, Bybjerg-Grauholm J, Bækvad-Hansen M, Cerrato F, Chambert K, Christensen JH, Churchhouse C, Dellenvall K, Demontis D, De Rubeis S, Devlin B, Djurovic S, Dumont AL, Goldstein JI, Hansen CS, Hauberg ME, Hollegaard MV, Hope S, Howrigan DP, Huang H, Hultman CM, Klei L, Maller J, Martin J, Martin AR, Moran JL, Nyegaard M, Nærland T, Palmer DS, Palotie A, Pedersen CB, Pedersen MG, dPoterba T, Poulsen JB, Pourcain BS, Qvist P, Rehnström K, Reichenberg A, Reichert J, Robinson EB, Roeder K, Roussos P, Saemundsen E, Sandin S, Satterstrom FK, Davey Smith G, Stefansson H, Steinberg S, Stevens CR, Sullivan PF, Turley P, Walters GB, Xu X; Autism Spectrum Disorder Working Group of the Psychiatric Genomics Consortium; BUPGEN; Major Depressive Disorder Working Group of the Psychiatric Genomics Consortium; 23andMe Research Team, Stefansson K, Geschwind DH, Nordentoft M, Hougaard DM, Werge T, Mors O, Mortensen PB, Neale BM, Daly MJ, Børglum AD (2019) Identification of common genetic risk variants for autism spectrum disorder. Nat Genet 51: 431–444. doi:10.1038/s41588-019-0344-8 ↩
- Satterstrom FK, Kosmicki JA, Wang J, Breen MS, De Rubeis S, An JY, Peng M, Collins R, Grove J, Klei L, Stevens C, Reichert J, Mulhern MS, Artomov M, Gerges S, Sheppard B, Xu X, Bhaduri A, Norman U, Brand H, Schwartz G, Nguyen R, Guerrero EE, Dias C Autism Sequencing Consortium; iPSYCH-Broad Consortium, Betancur C, Cook EH, Gallagher L, Gill M, Sutcliffe JS, Thurm A, Zwick ME, Børglum AD, State MW, Cicek AE, Talkowski ME, Cutler DJ, Devlin B, Sanders SJ, Roeder K, Daly MJ, Buxbaum JD (2020) Large-Scale Exome Sequencing Study Implicates Both Developmental and Functional Changes in the Neurobiology of Autism. Cell 180(3): 568-584. doi: 10.1016/j.cell.2019.12.036 ↩